MeilerLab members are frequently engrossed in the development and application of Rosetta protocols. The lab developed a so-called multi-state design approach which enables Rosetta protein design on multiple protein conformations simultaneously. Each protein conformation ideally represents one typical “state” of the protein. The design on multiple states as opposed to only one state, allows us to inform Rosetta about possible protein flexibility, which would otherwise be disregarded during single-state design (Figure 1a).
Expanding regular single state design with multiple states allows us to address more complex design goals. For example, antibodies can be designed to tightly bind multiple different antigens at the same time (positive design), or can be engineered to not bind to another epitope (negative design). Proteins designed this ways exhibit sequence motifs that tightly bind or disregard specific antigens. These sequence patterns enables us to engineer novel protein function: The promiscuous binding of different antigens.
Protein function and dynamics that arise from characteristic sequences are so tightly intertwined, that supporting motifs of similar proteins can be observed across different species and their evolutionary paths (homologous proteins).
Evolutionary preferences for sequence motifs can be extracted via co-evolutionary analysis of homologous protein sequences, and gives rise to large networks of sequence preferences that can span the whole protein. Evolutionary sequence analysis is an alternative approach to directly adding multiple structural states to Rosetta. The MeilerLab developed a Rosetta protocol that helps Rosetta to conserve these functionally crucial networks of sequence preferences. The resulting proteins are less artificial than otherwise computationally generated proteins. We also have shown, that protein function can be reliably conserved without the need of knowledge regarding the protein functions (Figure 1b).
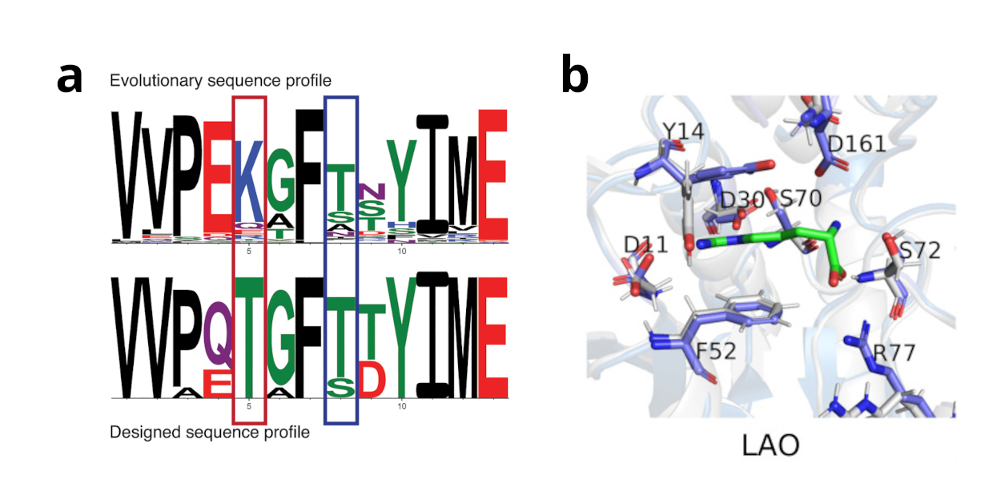
Figure 1: Rosetta multi-state design using the RECON protocol (a) is able to recapitulate certain sequence motifs which are important for all structural conformations used during design. These residues are more similar to those observed in evolution. Protein dynamic and functional information an also be integrated using co-evolutionary information using the ResCue protocol (b). Co-evolutionary information – if available – is capable to retain protein function and conserve functionally important
