Protein-ligand interaction studies are important for understanding the mechanisms of biological regulation, and they provide a theoretical basis for the design and discovery of new drug targets. To predict the Protein-Ligand interactions (PLIs), we dock the ligand to the target protein with RosettaLigand docking application. The predicted PIIs are then characterized by breaking down the Rosetta interface energy scores into per-residue level to identify potential lost-of-functions and gain-of-function mutants, which can further validate the predicted PLIs.
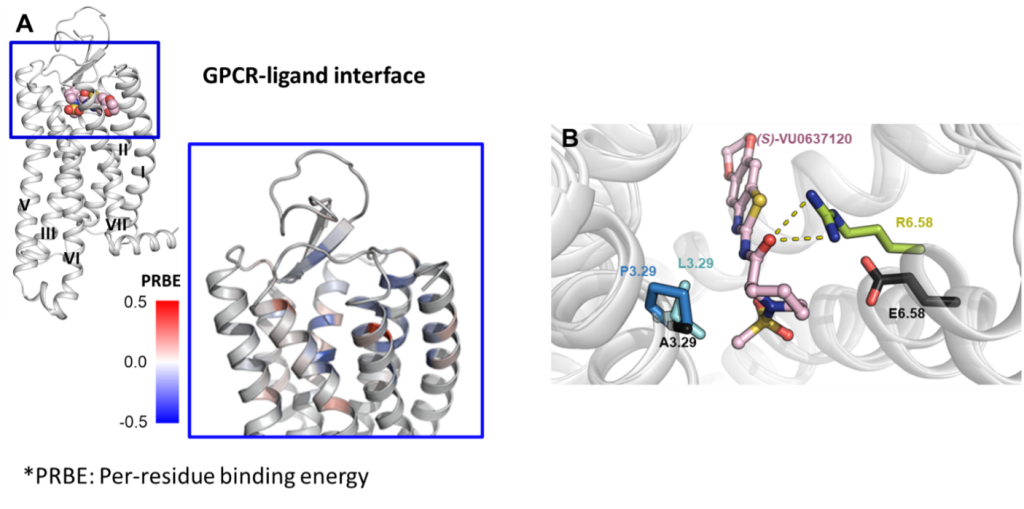
Figure 1: Protein-Ligand Interactions
(A) Mapping the Rosetta per-residue binding energy onto Y4 receptor to characterize its PIIs to (S)-VU0637120, a Y4 partial NAM (shown in pink spheres – upper left figure). The binding pocket residues are colored according to the magnitude of binding energy such that blue is more negative (more favorable), and red is more positive (less favorable) (lower right figure).
(B) Illustration of the GOF mutants (E6.58R, A3.29L, and A3.29P) in the docking model. Residues are numbered according to the nomenclature of Ballosteros-Weinstein.
Bibliography
[1]
Zimmermann A, Vu O, Brüser A, Sliwoski G, Marnett LJ, Meiler J, Schöneberg T. Mapping the Binding Sites of UDP and Prostaglandin E2 Glyceryl Ester in the Nucleotide Receptor P2Y6. ChemMedChem. 2022 Jan 14:e202100683.
[2]
Schüß C, Vu O, Schubert M, Du Y, Mishra NM, Tough IR, Stichel J, Weaver CD, Emmitte KA, Cox HM, Meiler J, Beck-Sickinger AG. Highly Selective Y4 Receptor Antagonist Binds in an Allosteric Binding Pocket. J Med Chem. 2021 Mar 11;64(5):2801-2814.
[3]
Moretti R, Bender BJ, Allison B, Meiler J. Rosetta and the Design of Ligand Binding Sites. Methods Mol Biol. 2016;1414:47-62.
