Cholesterol (CLR) has been shown to alter integral membrane protein (IMP) function. However, understanding the molecular mechanisms of this process is complicated by limited and conflicting structural data. Specifically, in co-crystal structures of CLR-IMP complexes, it is difficult to distinguish a specific interaction with biological relevance from a nonspecific association captured due to the crystallization process. It is a daunting challenge to disentangle CLR’s dual roles as a modulator of IMP function through direct binding and as an indirect effector of membrane fluidity. Only recently have studies delved into characterizing specific interactions. For example, CLR binding to the endothelial Kir2.1 channel impairs flow-induced vasodilation and augments atherosclerosis development. We use evolutionary computations In conjunction with previous structural data to detect likely-specific from nonspecific interaction sites. Computational IMP design has the potential to provide a general, complementary approach for engineering CLR recognition in which the designed IMP features a specificity that can be rationally programmed.
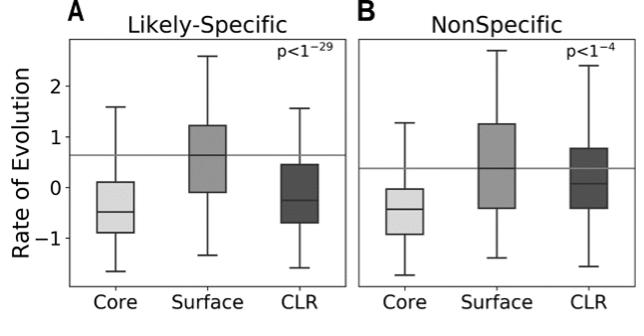
Figure: Likely-specific cholesterol binding sites are more conserved (negative rate of evolution)
