| Date | 2024-01-02 |
|---|---|
| Case | SampleCase |
| Gene | RTEL1 |
| Protein | Q9NZ71-8 |
| Variant | G546D |
MusiteDeep (PTMs) within 8.0A of G546
| PTM Residue | PTM Type | Probability |
|---|---|---|
| 543 | Ubiquitination | 0.6 |
| 586 | N6-acetyllysine | 0.6 |
Structure Summary
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| swiss | Q9NZ71-8_3_755_7o75.1.A | A | 7o75.1.A | 26.56 | 1.0 | 3 | 755 |
Q9NZ71-8_3_755_7o75.1.A.A monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| swiss | Q9NZ71-8_3_755_7o75.1.A | A | 7o75.1.A | 26.56 | 1.0 | 3 | 755 |
clinvar Results - Q9NZ71-8_3_755_7o75.1.A.A
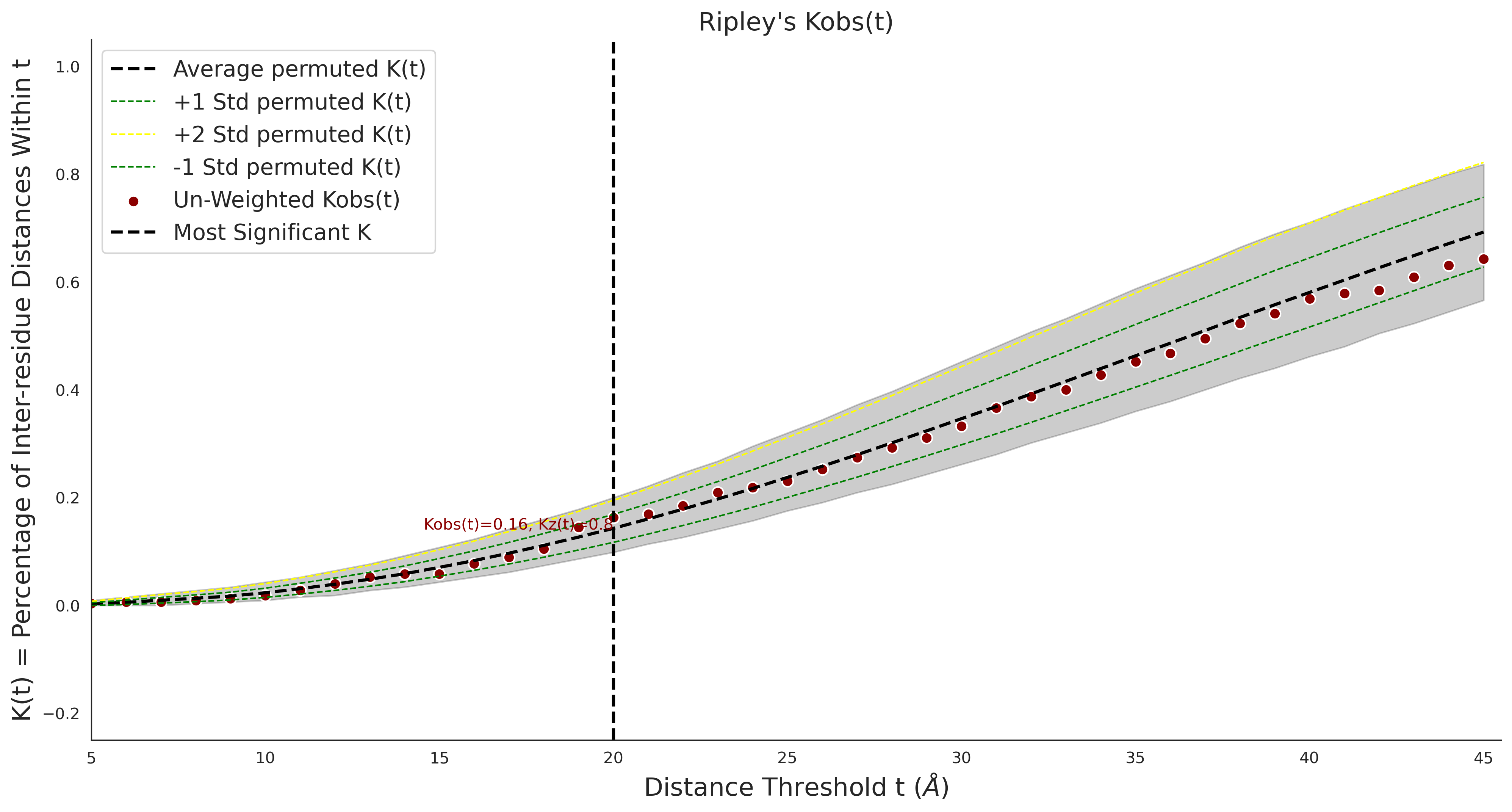
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| 0.00 | 0.00 | 1.43 | 13.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.10 |
COSMIC Results - Q9NZ71-8_3_755_7o75.1.A.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| 0.00 | 0.00 | 0.78 | 20.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 |
Location of G546D in Q9NZ71-8_3_755_7o75.1.A.A (NGL Viewer)
Variant Distributions
gnomAD38
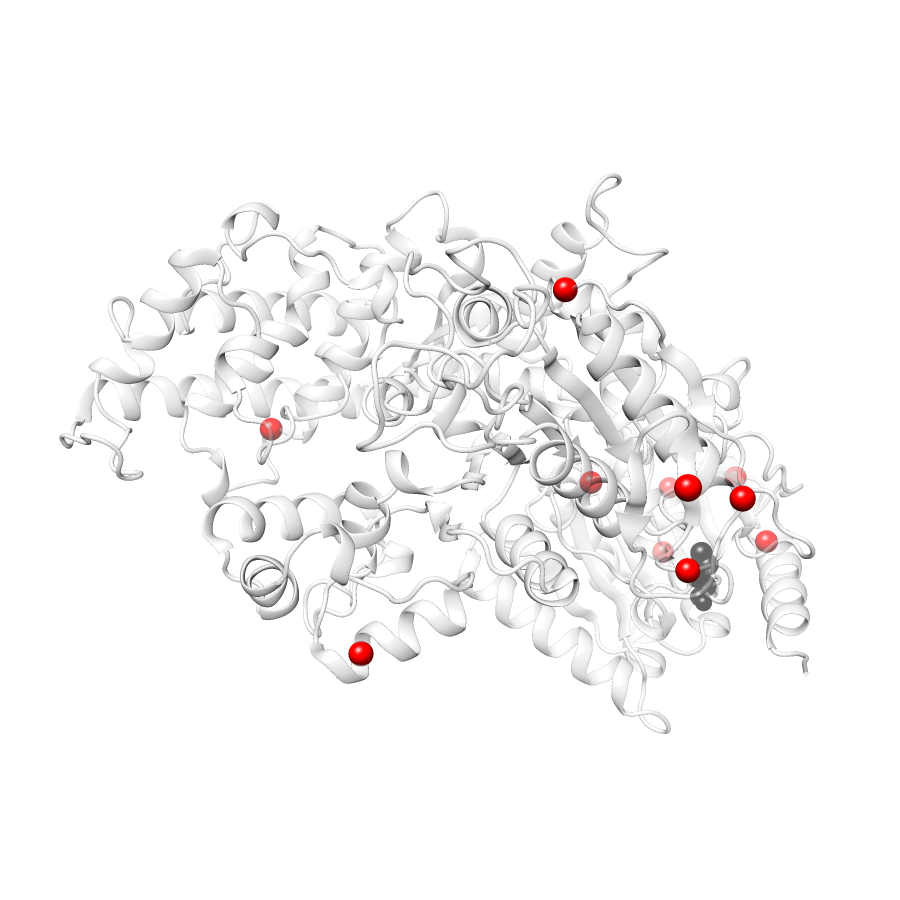
11 clinvar
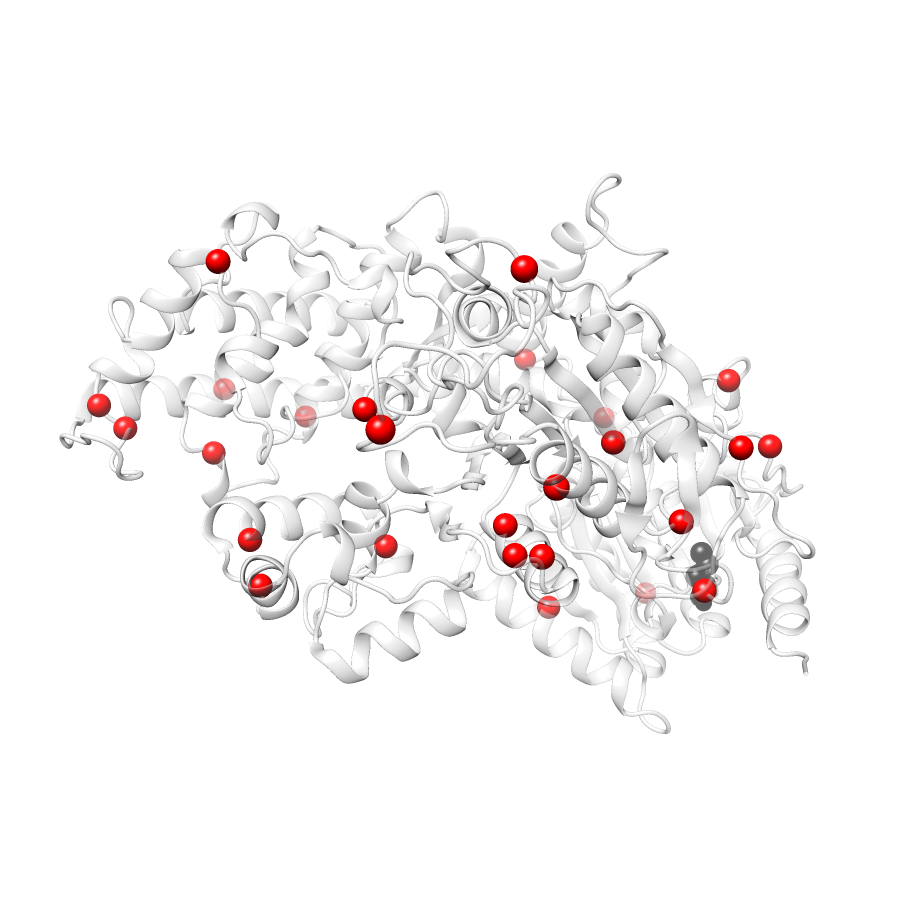
26 COSMIC
Ripley's Univariate K
gnomAD38
Ripley's Univariate K graphic was not created for gnomAD38clinvar
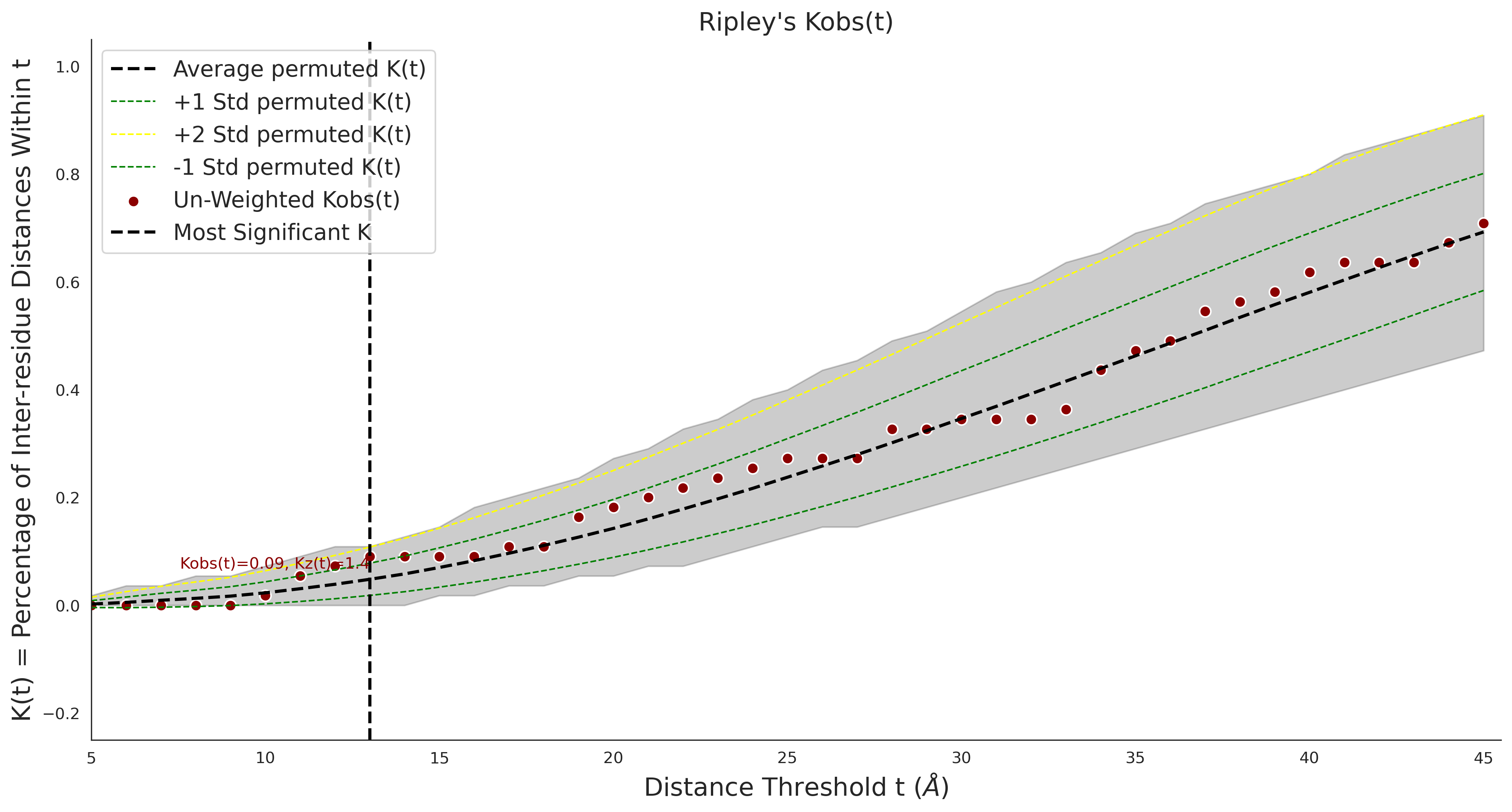
COSMIC