| Date | 2024-01-02 |
|---|---|
| Case | SampleCase |
| Gene | ONECUT1 |
| Protein | Q9UBC0 |
| Variant | R269G |
R269 Rate4Site Score ENST00000305901.7
| Rate4Site Score | QQ-Interval | Std | MSA | |
|---|---|---|---|---|
| Normalized: | -0.48 | -0.63, -0.27 | 0.26 | 96.00/100 |
No MusiteDeep (PTMs) predicted within 8.0A of R269
ScanNet Prediction of R269 participation in a PPI
| Binding Site Probability |
|---|
| 0.3 |
Structure Summary
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| alphafold | AF-Q9UBC0-F1 | A | 0.0 | 1.0 | 60.92 | 42.38 | 1 | 465 | -2.92 | -1.97 | 0.04 | 0.706 |
AF-Q9UBC0-F1.A monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| alphafold | AF-Q9UBC0-F1 | A | 0.0 | 1.0 | 60.92 | 42.38 | 1 | 465 | -2.92 | -1.97 | 0.04 | 0.706 |
clinvar Results - AF-Q9UBC0-F1.A
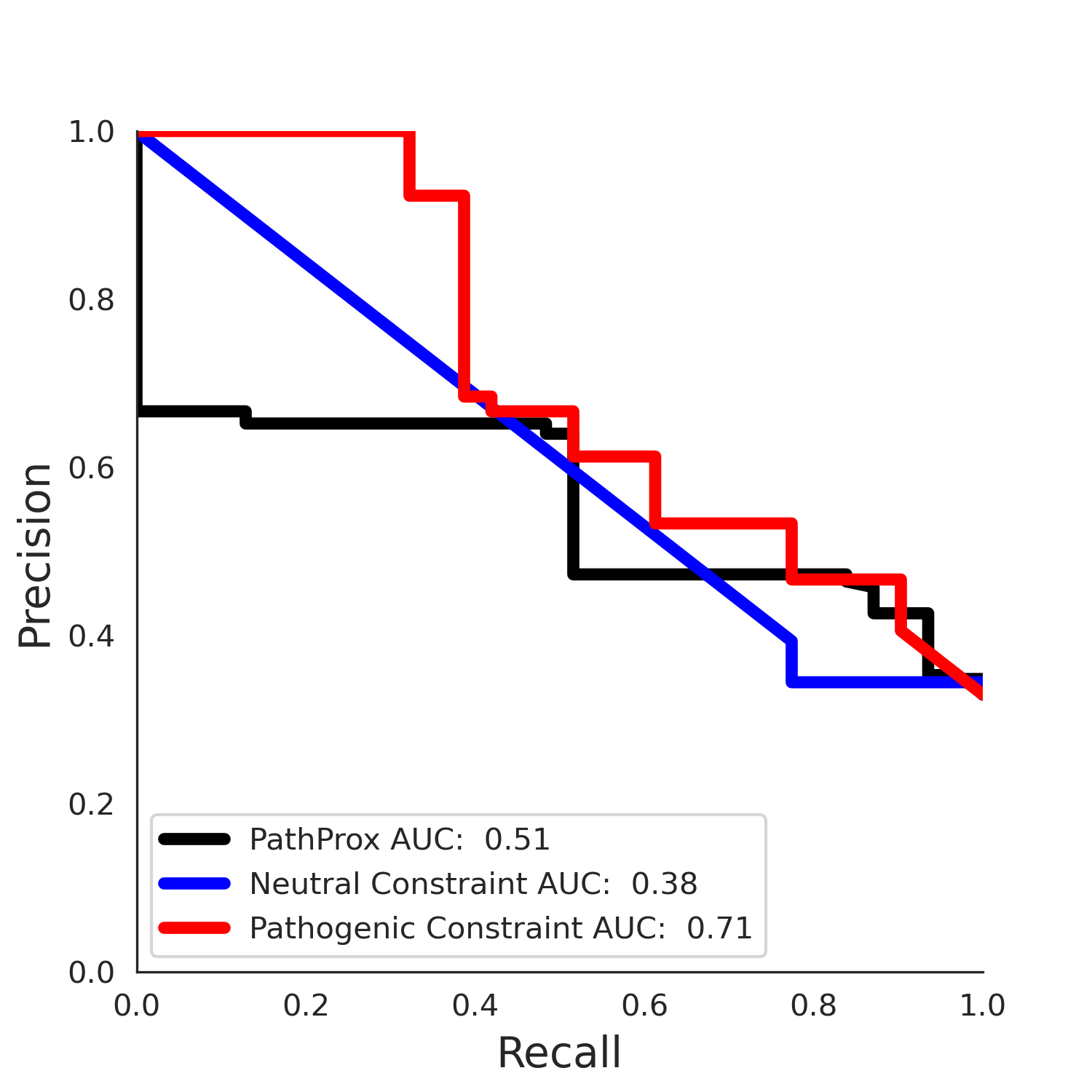
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| 3.05 | 5.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 |
COSMIC Results - AF-Q9UBC0-F1.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
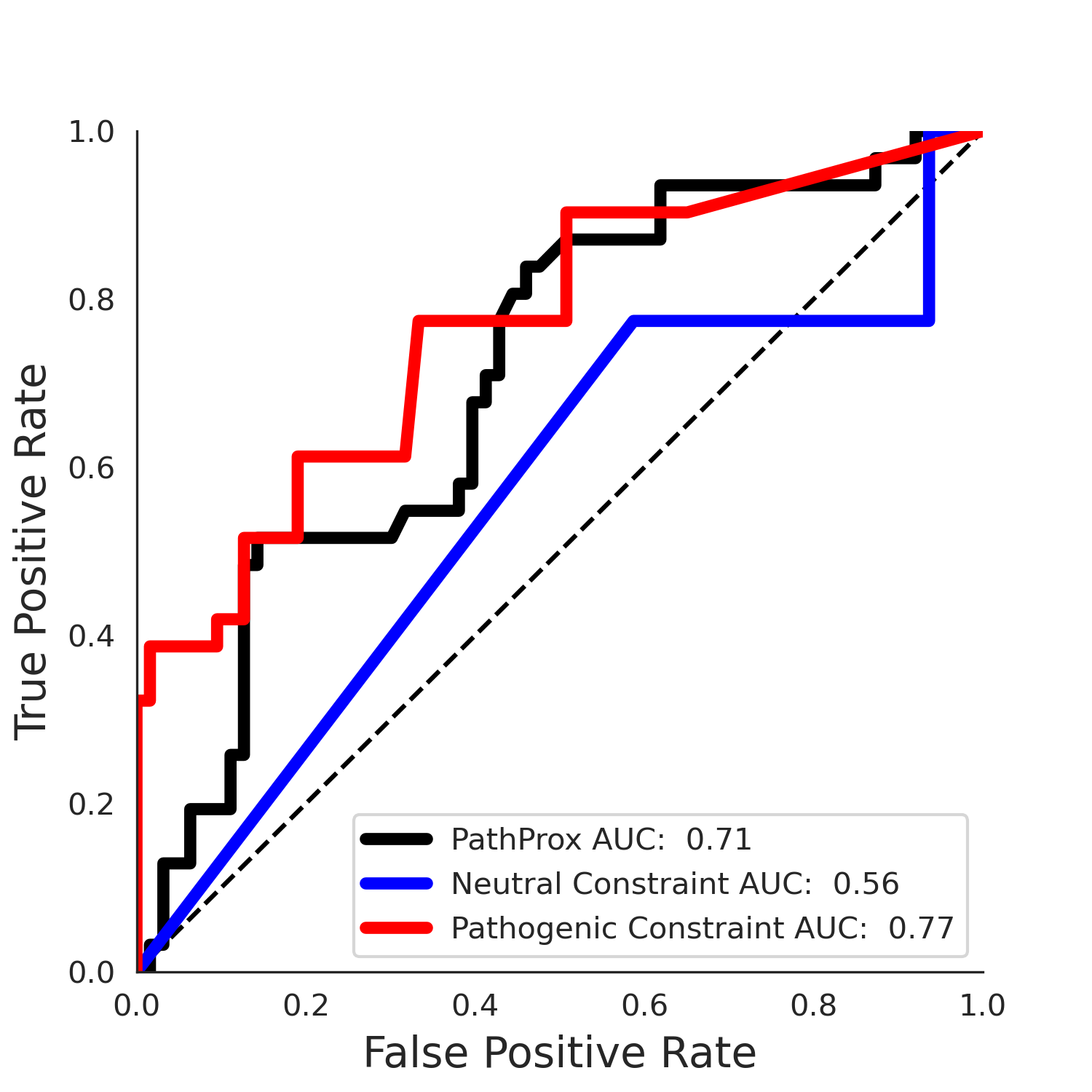
| 3.55 | 5.00 | 2.35 | 29.00 | 0.77 | 0.71 | 0.04 | 1.00 | 0.01 |
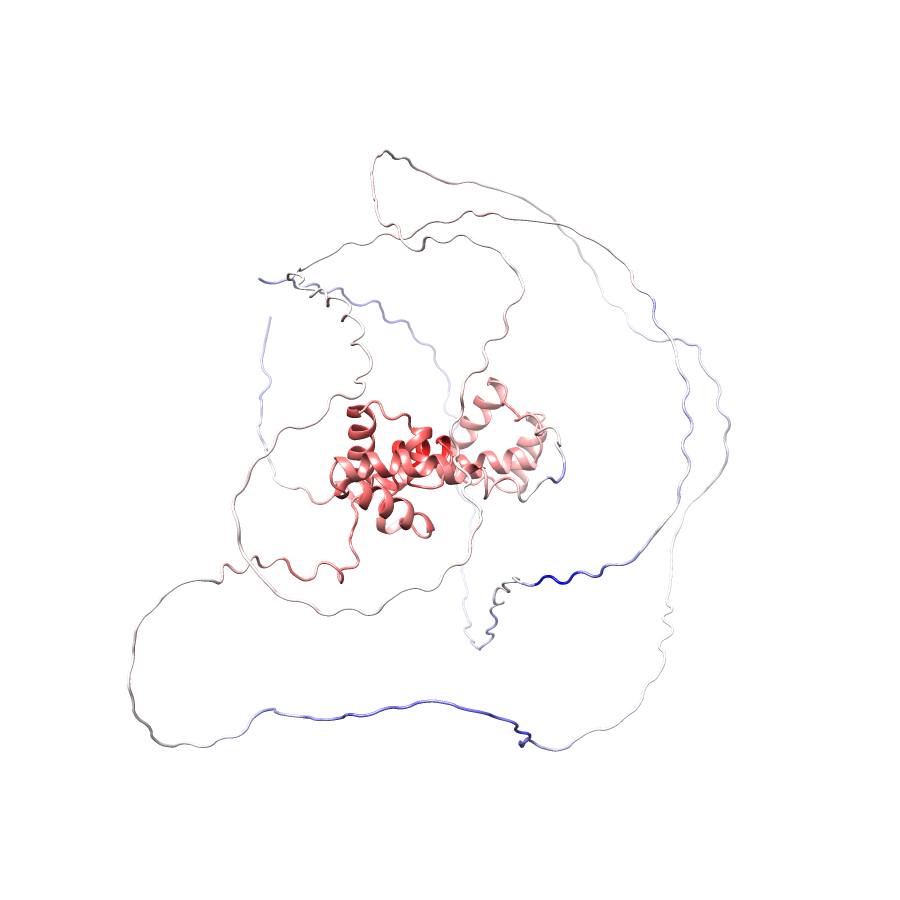
Location of R269G in AF-Q9UBC0-F1.A (NGL Viewer)
Variant Distributions
63 gnomAD38
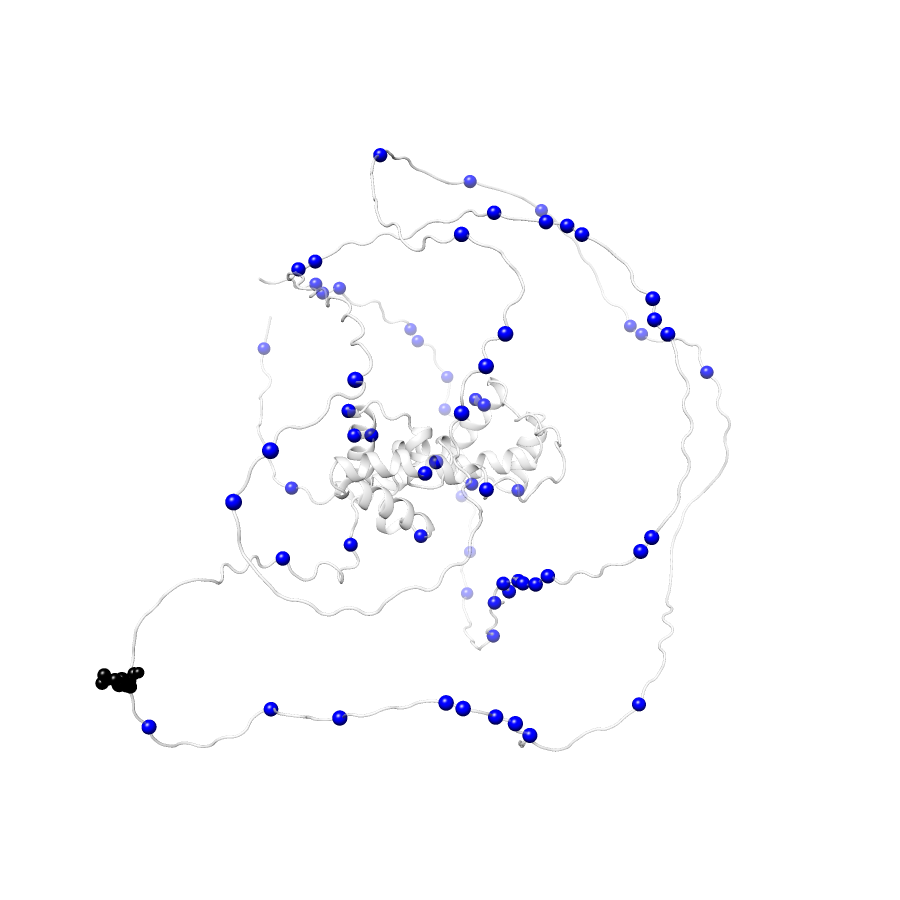
clinvar
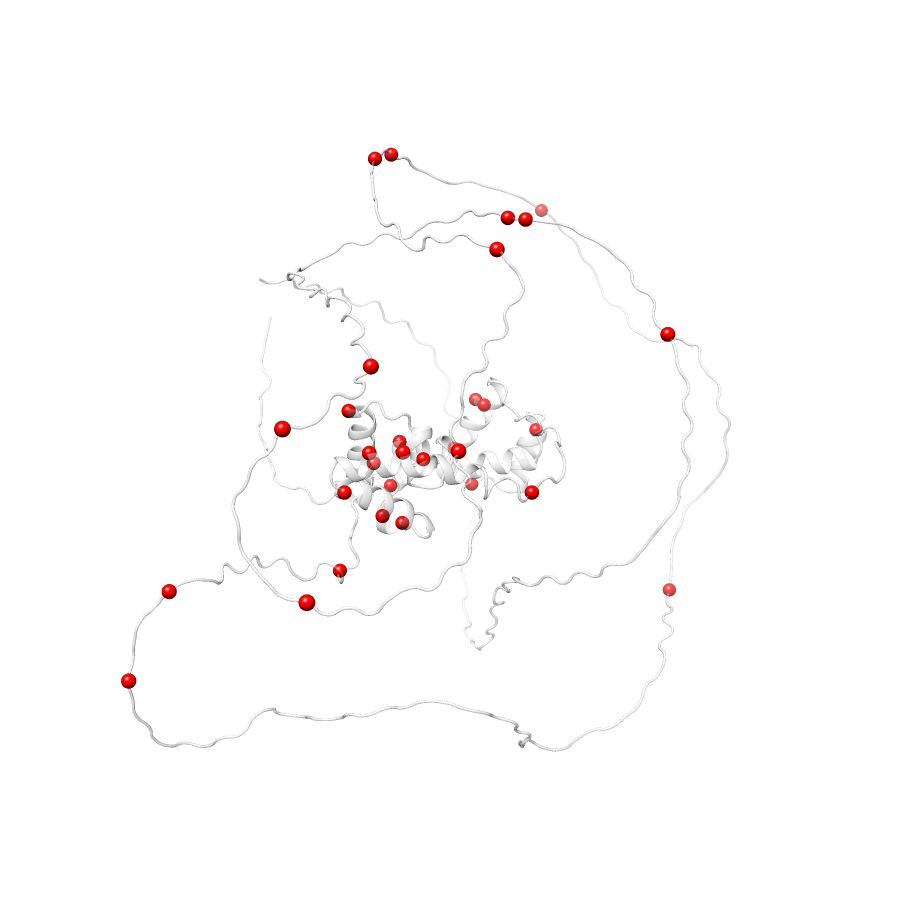
No clinvar Variants map to this structure31 COSMIC
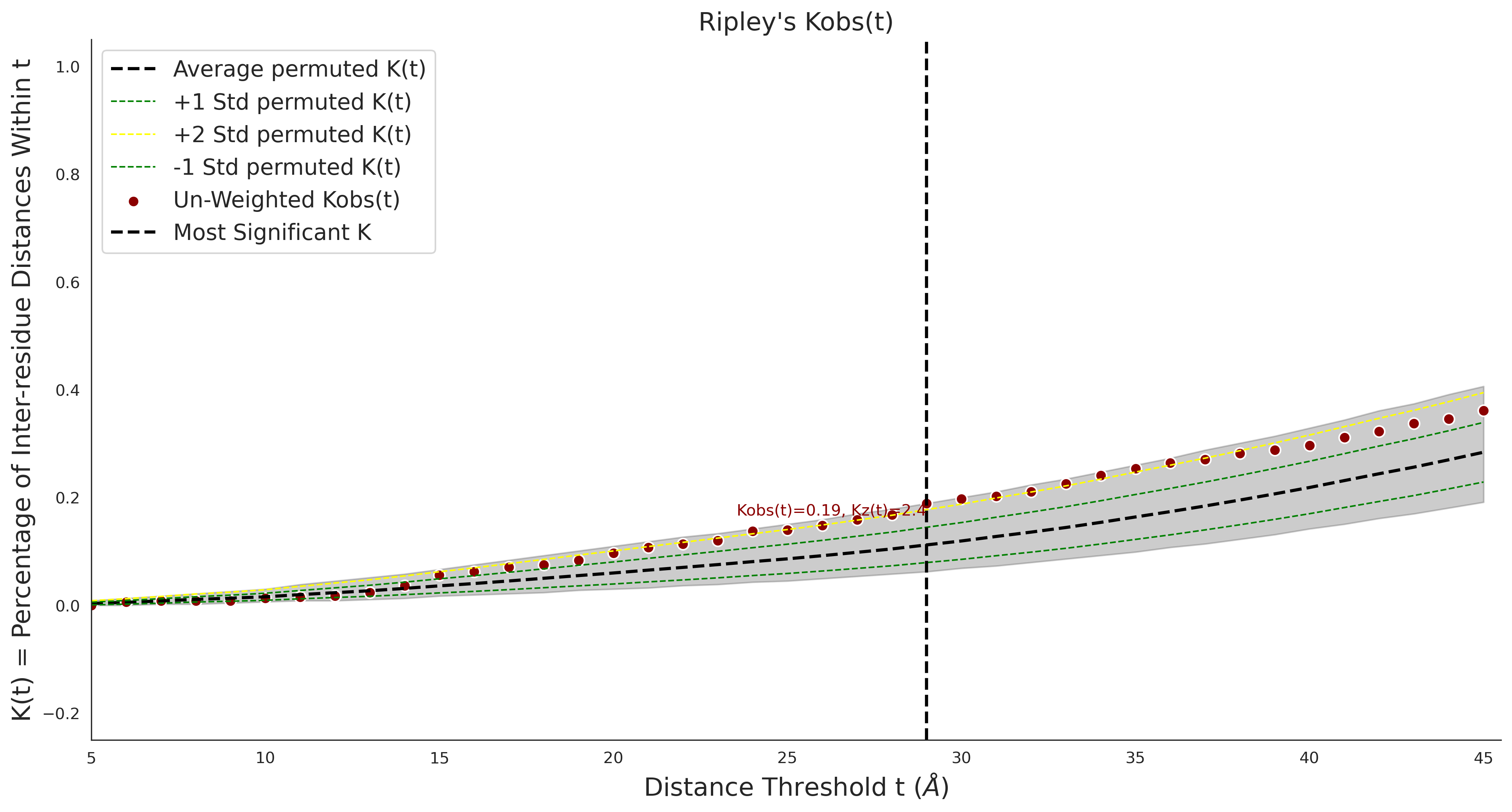
Ripley's Univariate K
gnomAD38
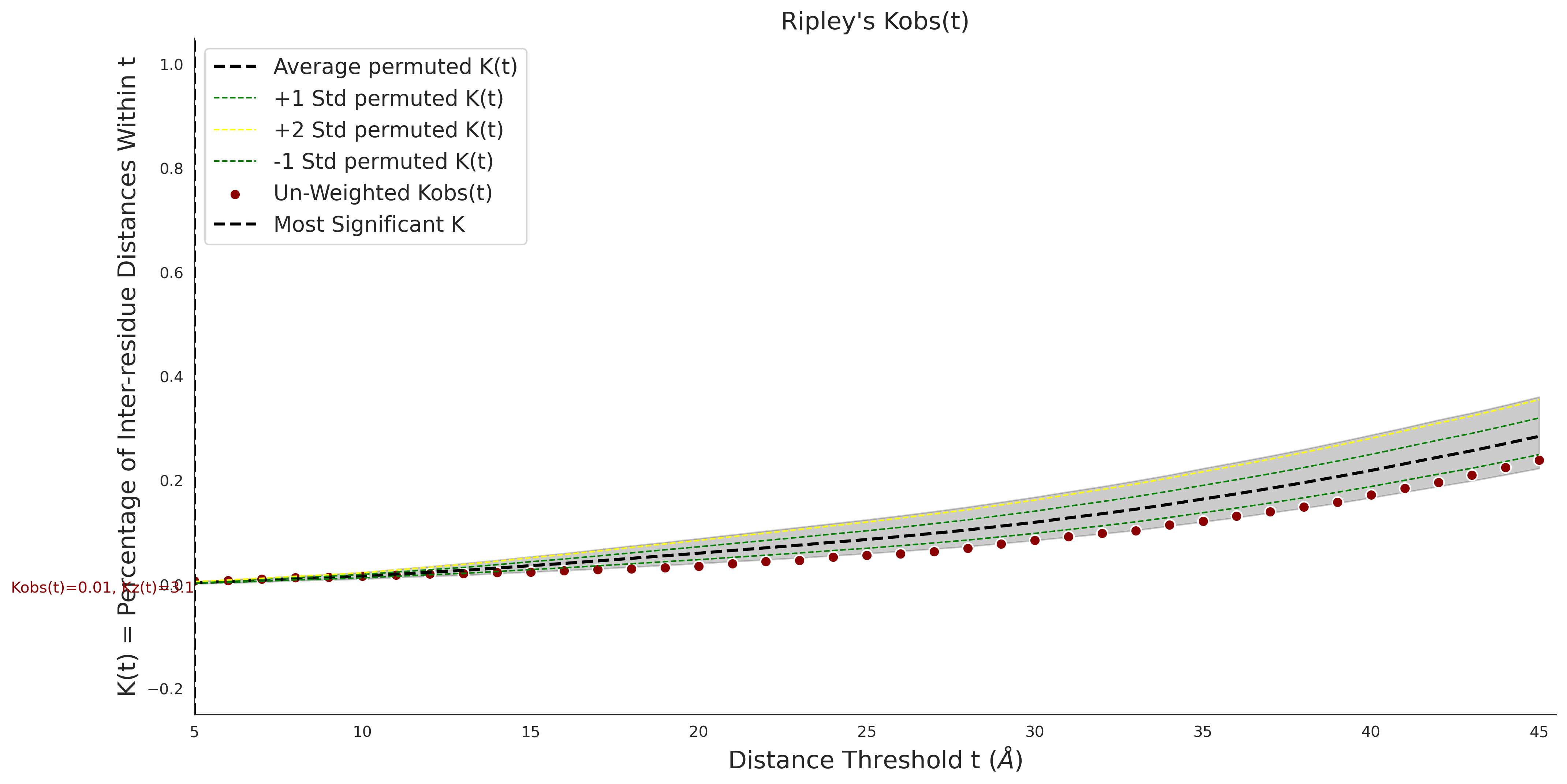
clinvar
Ripley's Univariate K graphic was not created for clinvarCOSMIC
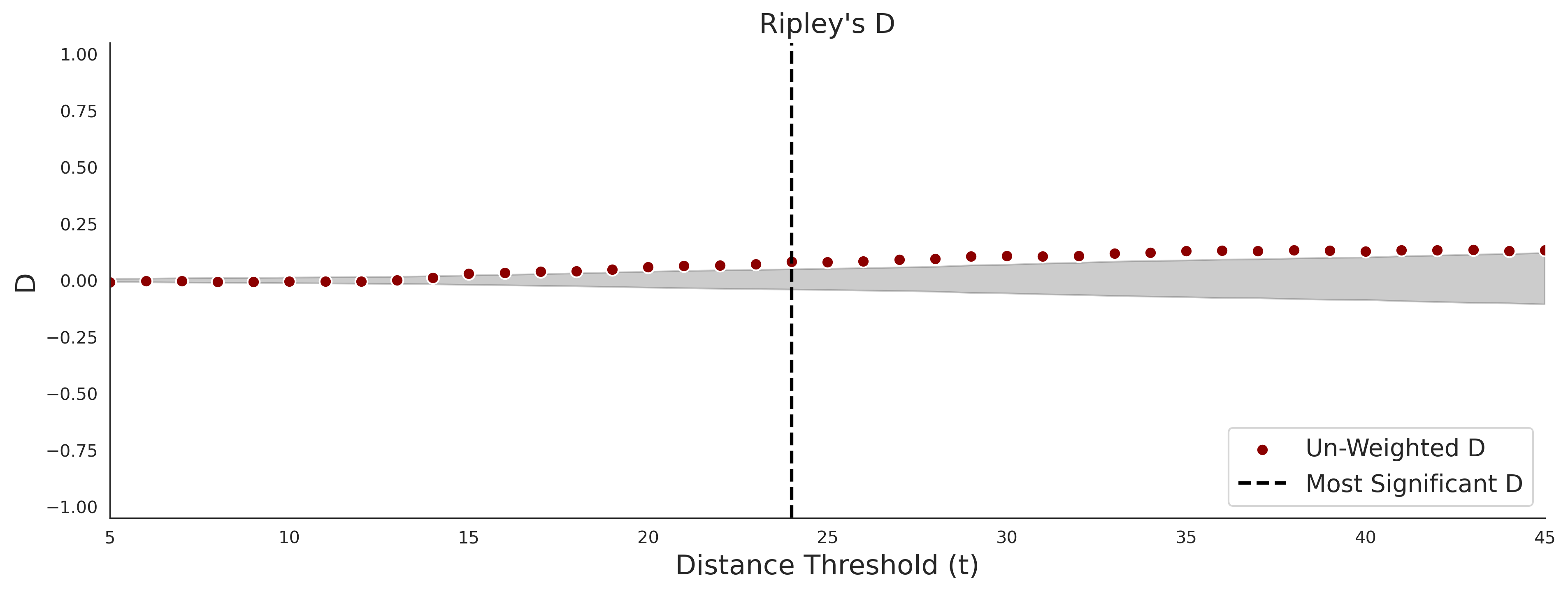
Ripley's Bivariate D
clinvar - gnomAD38
clinvar-gnomAD38 Bivariate D plot was not createdCOSMIC - gnomAD38
PathProx (clinvar - gnomAD38) Results
PathProx
No graphic was createdPathProx clinvar ROC
No graphic was createdPathProx clinvar PR
No graphic was createdPathProx (COSMIC - gnomAD38) Results
PathProx
PathProx COSMIC ROC
PathProx COSMIC PR