| Date | 2024-01-02 |
|---|---|
| Case | SampleCase |
| Gene | AP3D1 |
| Protein | O14617-5 |
| Variant | V699M |
MusiteDeep (PTMs) within 8.0A of V699
| PTM Residue | PTM Type | Probability |
|---|---|---|
| 696 | Phosphothreonine | 0.7 |
Structure Summary
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| modbase | ENSP00000495274.1_2 | A | 4pjw | 14.0 | 1.0 | 313 | 1019 | 0.03 | 0.299 |
ENSP00000495274.1_2.A monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| modbase | ENSP00000495274.1_2 | A | 4pjw | 14.0 | 1.0 | 313 | 1019 | 0.03 | 0.299 |
clinvar Results - ENSP00000495274.1_2.A
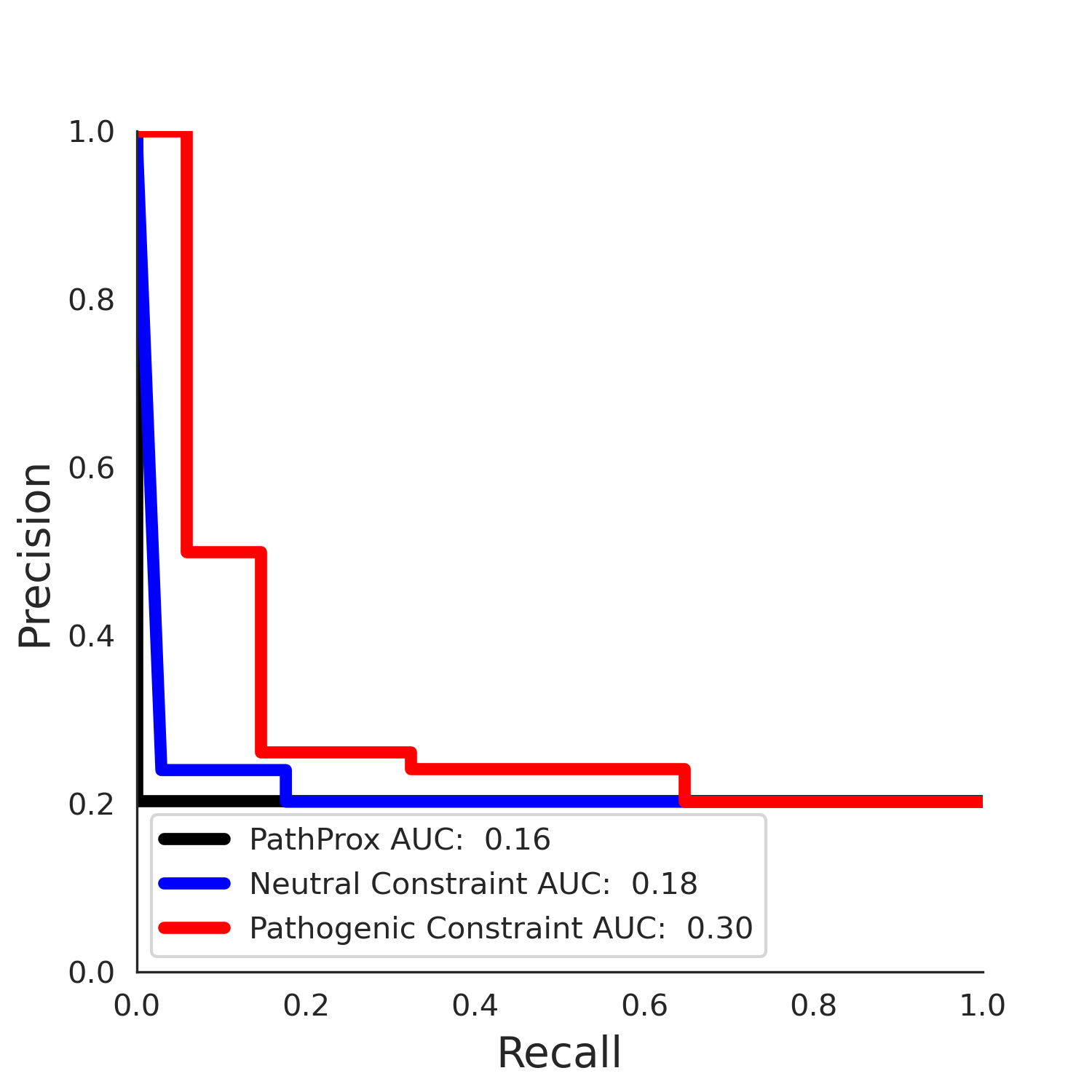
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -2.28 | 11.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.97 | 0.00 |
COSMIC Results - ENSP00000495274.1_2.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
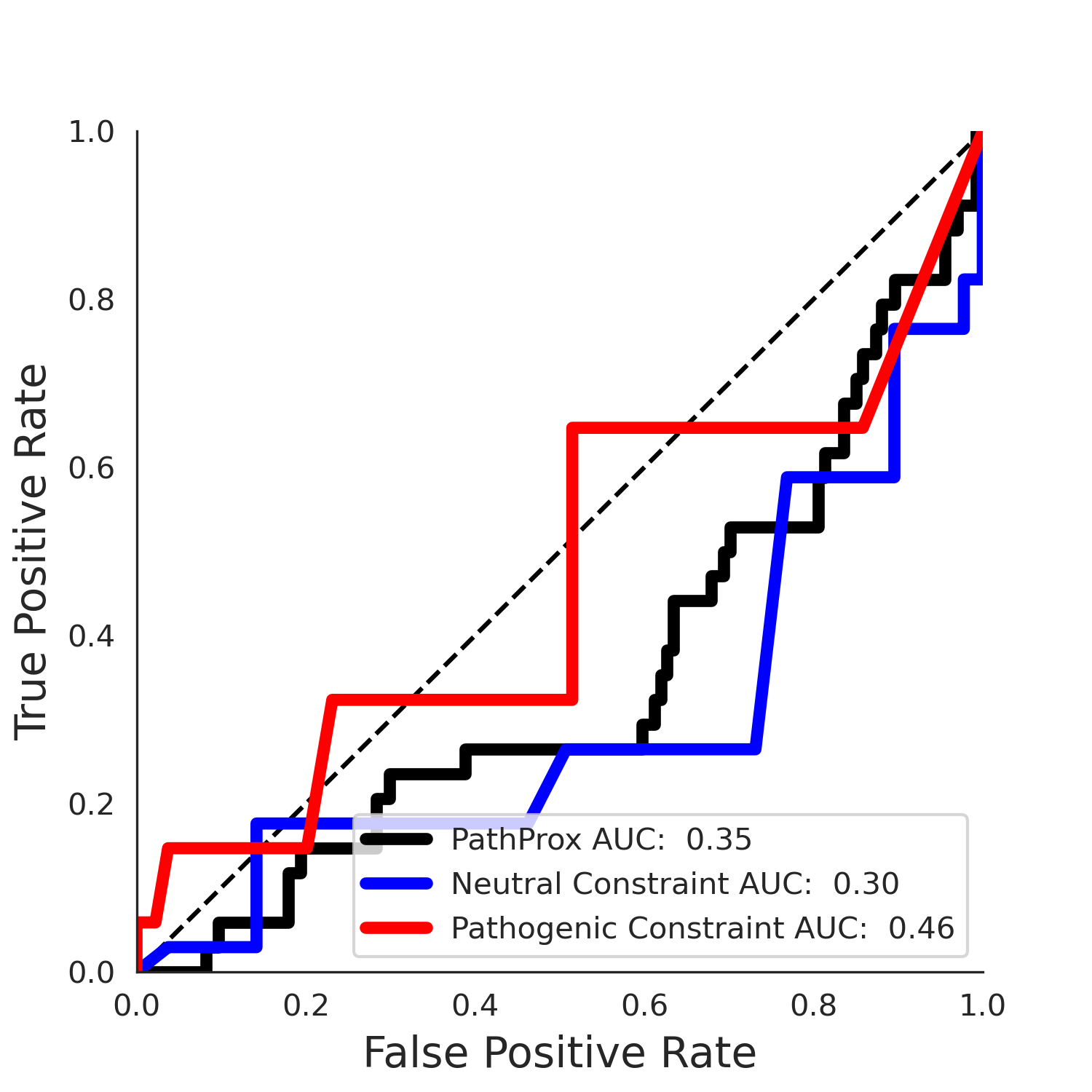
| -2.33 | 11.00 | -1.90 | 14.00 | 0.46 | 0.30 | 0.03 | 0.98 | 0.03 |
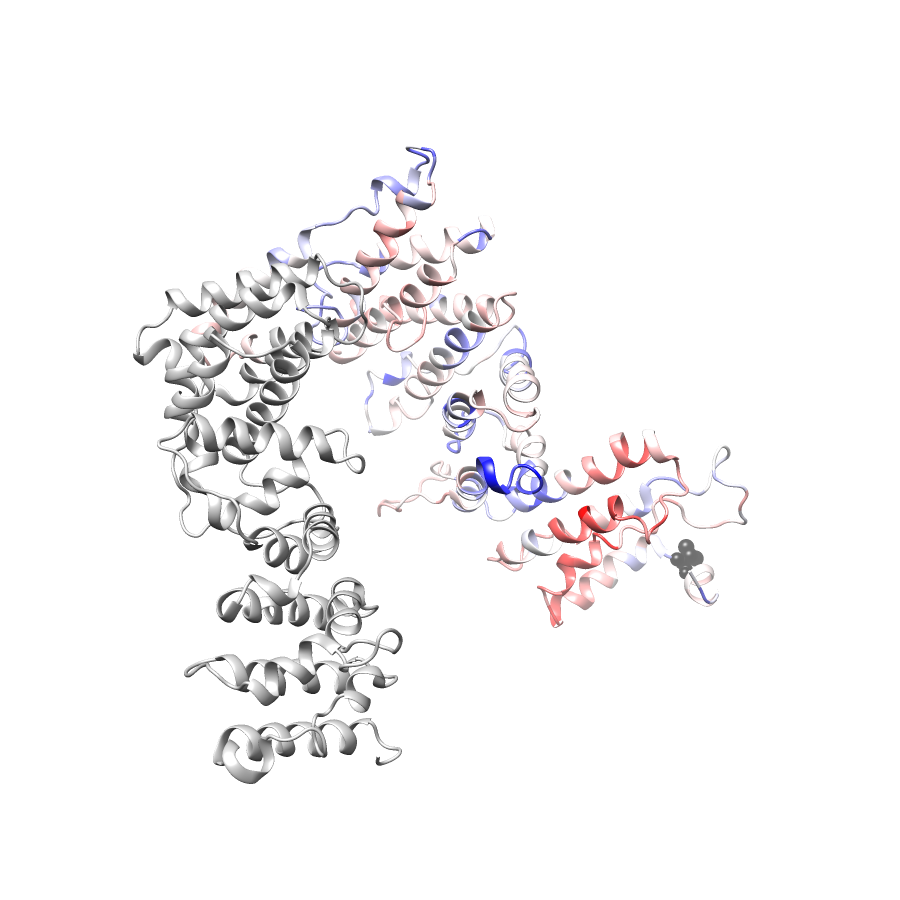
Location of V699M in ENSP00000495274.1_2.A (NGL Viewer)
Variant Distributions
134 gnomAD38
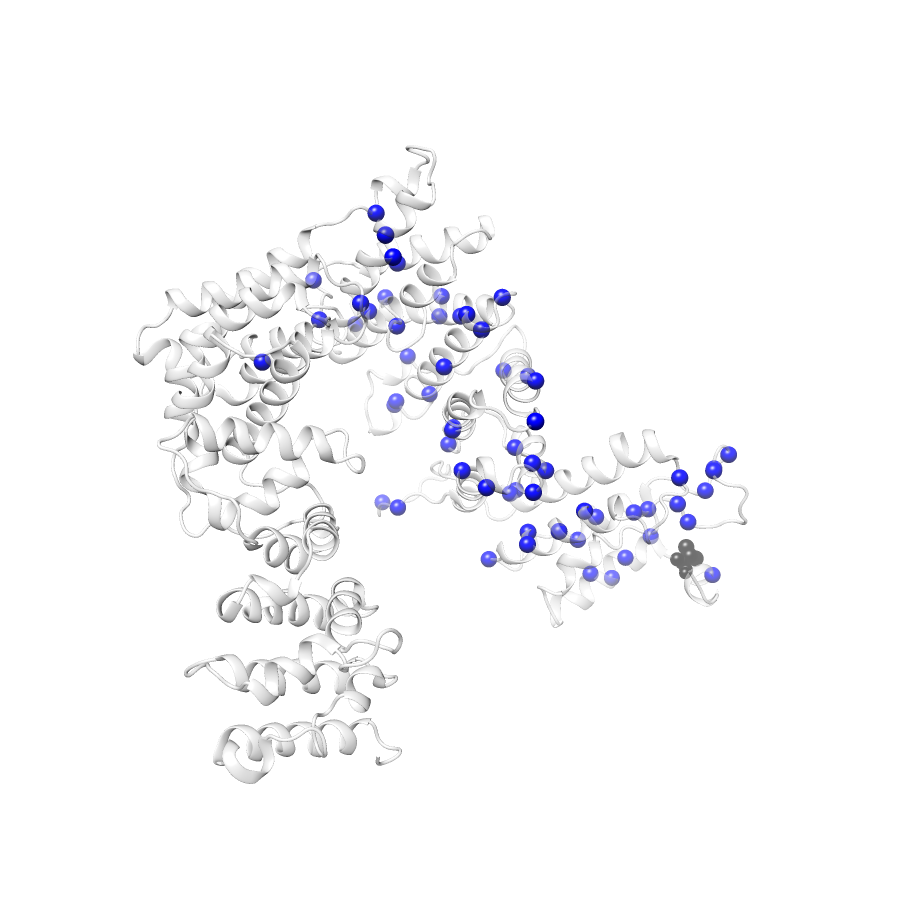
clinvar
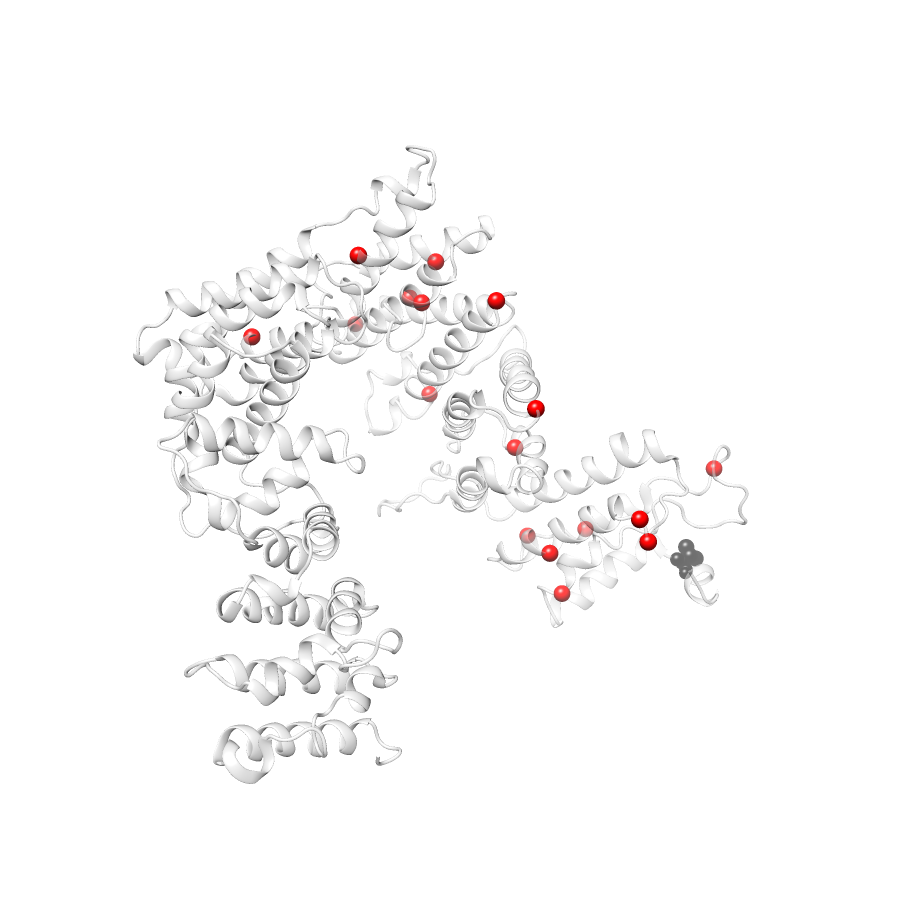
No clinvar Variants map to this structure34 COSMIC
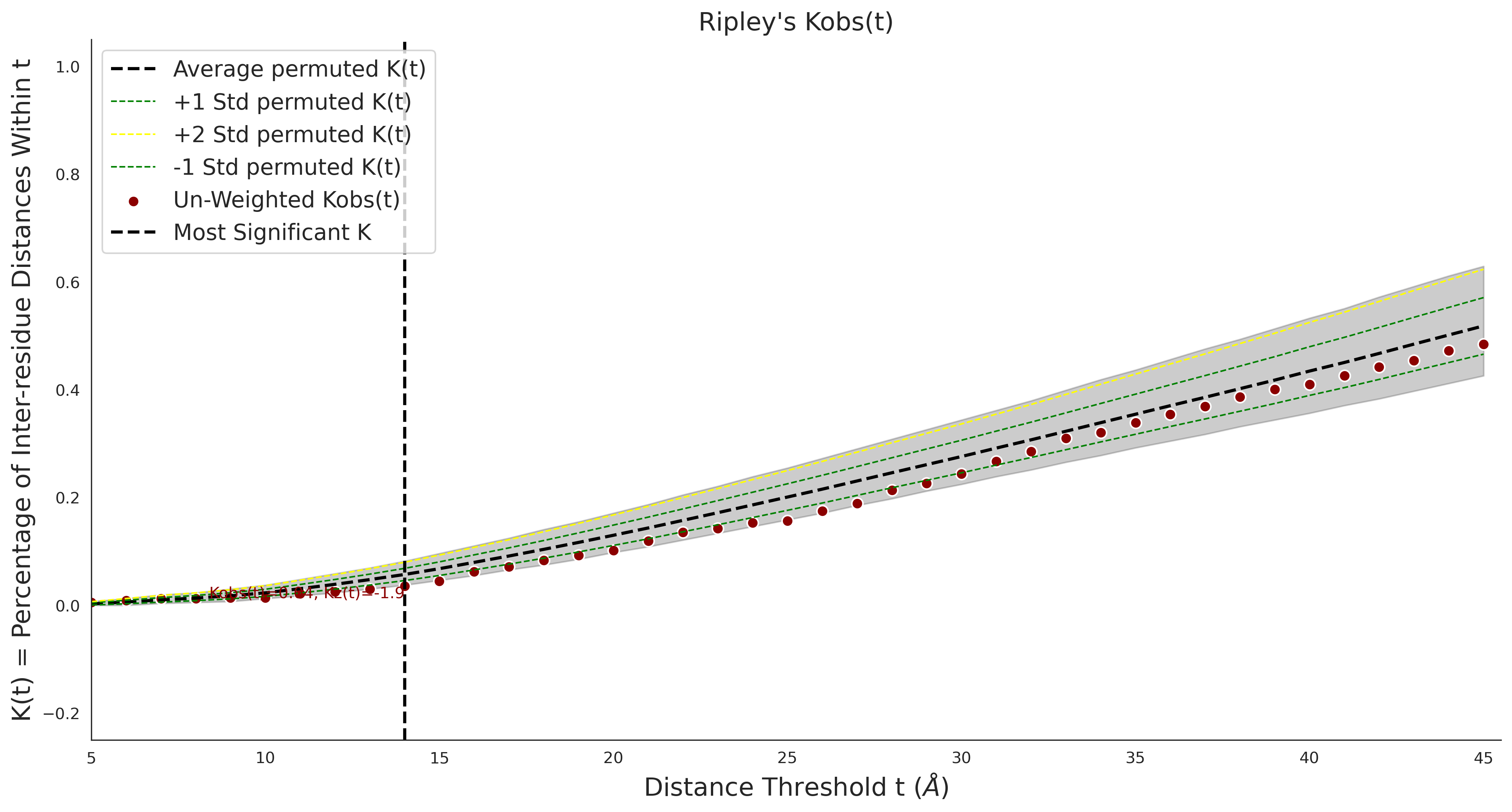
Ripley's Univariate K
gnomAD38
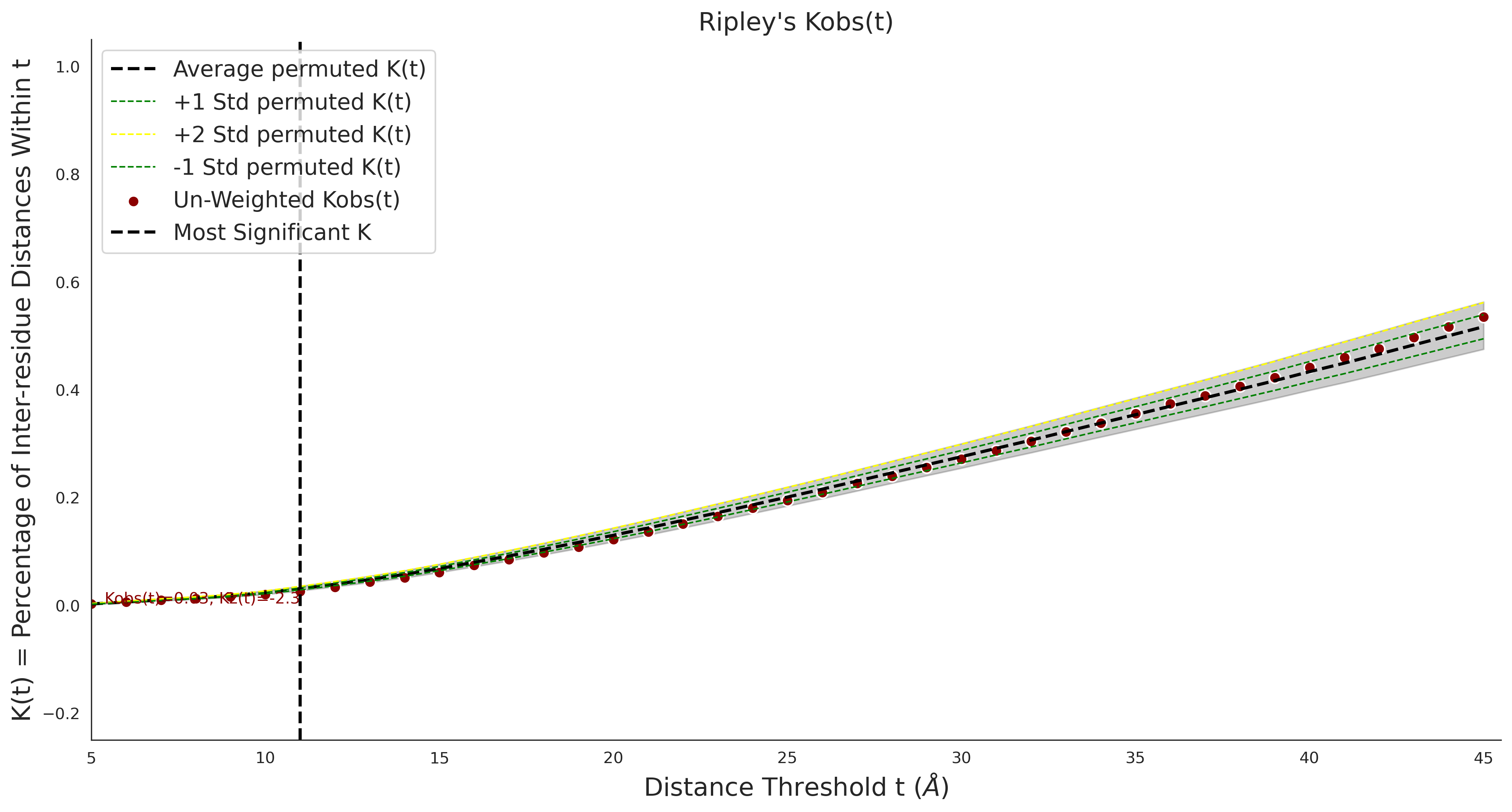
clinvar
Ripley's Univariate K graphic was not created for clinvarCOSMIC
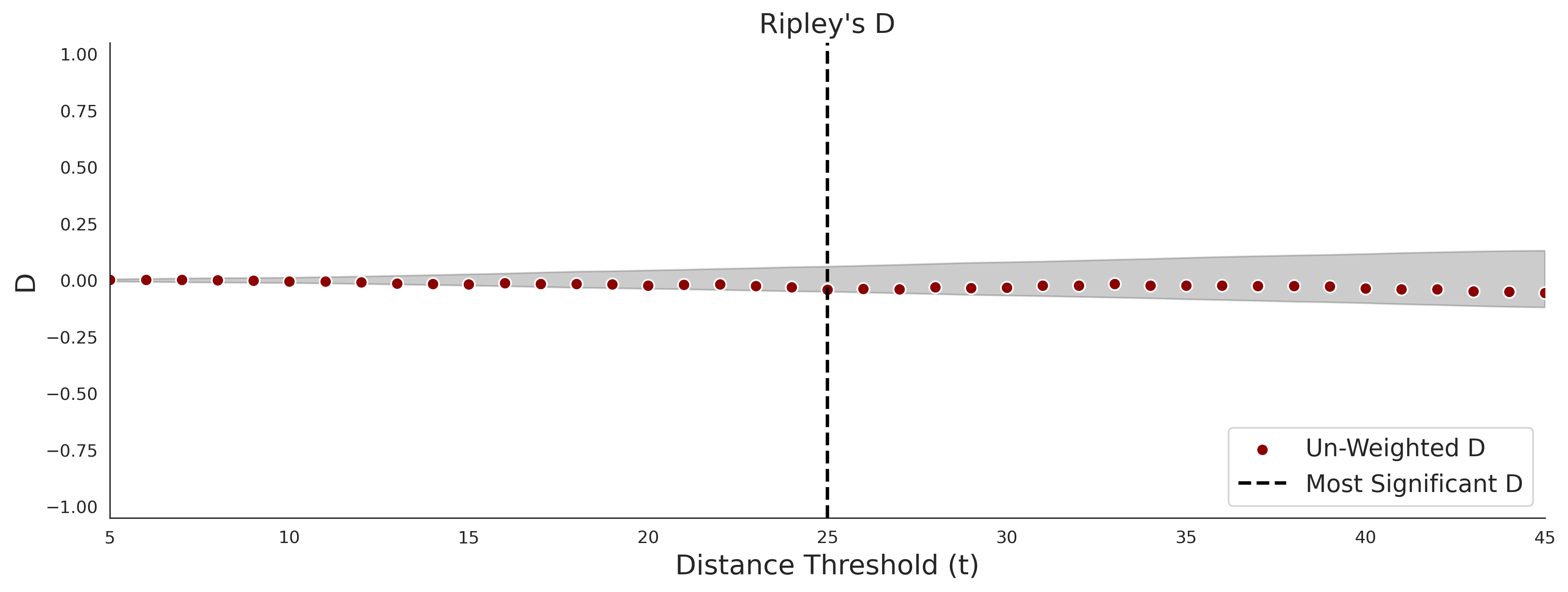
Ripley's Bivariate D
clinvar - gnomAD38
clinvar-gnomAD38 Bivariate D plot was not createdCOSMIC - gnomAD38
PathProx (clinvar - gnomAD38) Results
PathProx
No graphic was createdPathProx clinvar ROC
No graphic was createdPathProx clinvar PR
No graphic was createdPathProx (COSMIC - gnomAD38) Results
PathProx
PathProx COSMIC ROC
PathProx COSMIC PR