| Date | 2024-01-02 |
|---|---|
| Case | SampleCase |
| Gene | ALAS2 |
| Protein | P22557-2 |
| Variant | A245T |
No MusiteDeep (PTMs) predicted within 8.0A of A245
Structure Summary
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| swiss | P22557-2_106_541_6hrh.1.A | B | 6hrh.1.A | 99.78 | 1.0 | 106 | 541 | 23.22 | 9.71 | 0.46 | 0.966 | 0.03 | 0.527 | ||||
| swiss | P22557-2_106_541_6hrh.1.A | B | 6hrh.1.A | 99.78 | 1.0 | 106 | 541 | 0.06 | 0.797 | 0.02 | 0.494 | ||||||
| modbase | ENSP00000337131.4_1 | A | 5qr1 | 99.0 | 1.0 | 103 | 541 | 17.06 | 11.32 | 0.5 | 0.958 | -0.01 | 0.471 | ||||
| modbase | ENSP00000337131.4_2 | A | 5txt | 37.0 | 1.0 | 106 | 510 | 0.37 | 0.908 | 0.03 | 0.498 | ||||||
| modbase | ENSP00000337131.4_3 | A | 2bwn | 50.0 | 1.0 | 107 | 503 | 2.3 | 2.97 | 0.29 | 0.874 | -0.02 | 0.479 | ||||
| X-RAY | 5qqr | A | 1.46 | 1.0 | 106 | 541 | -0.03 | 0.784 | -0.02 | 0.507 | |||||||
| X-RAY | 5qqr | B | 1.46 | 1.0 | 106 | 541 | 23.86 | 8.74 | 0.31 | 0.954 | -0.03 | 0.53 |
P22557-2_106_541_6hrh.1.A.B monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| swiss | P22557-2_106_541_6hrh.1.A | B | 6hrh.1.A | 99.78 | 1.0 | 106 | 541 | 23.22 | 9.71 | 0.46 | 0.966 | 0.03 | 0.527 |
clinvar Results - P22557-2_106_541_6hrh.1.A.B
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
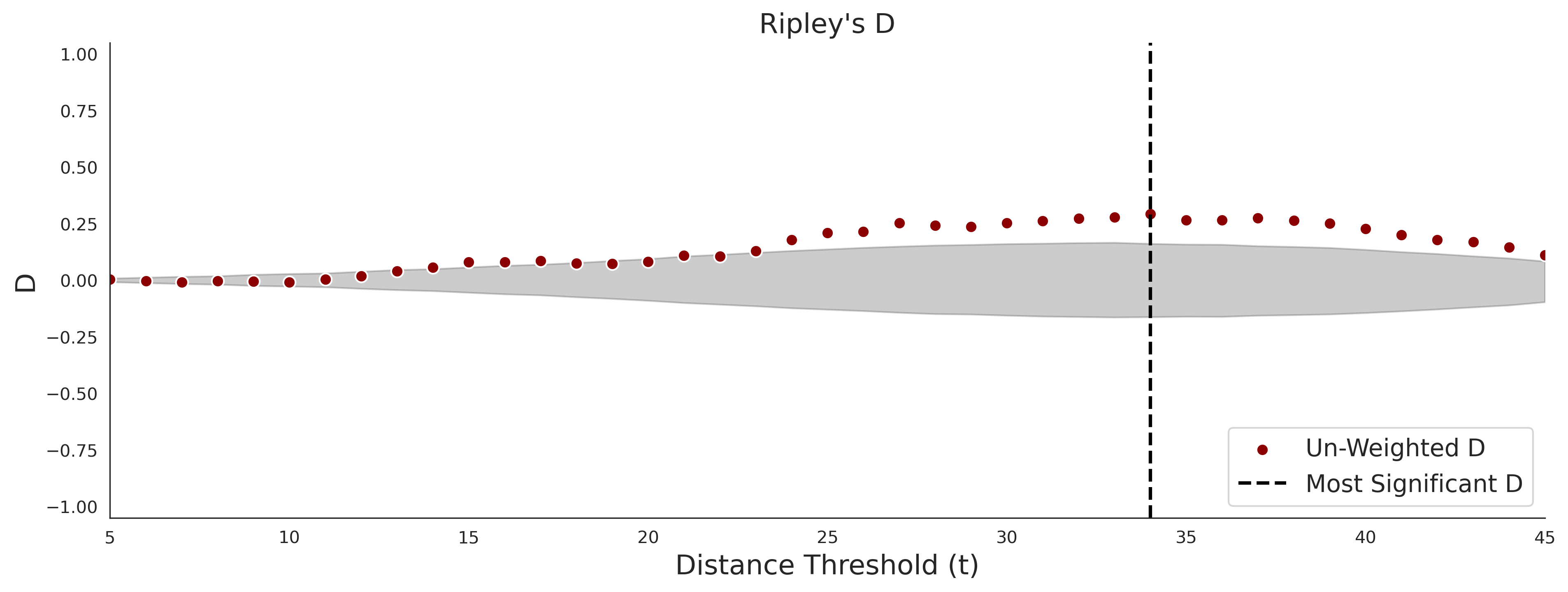
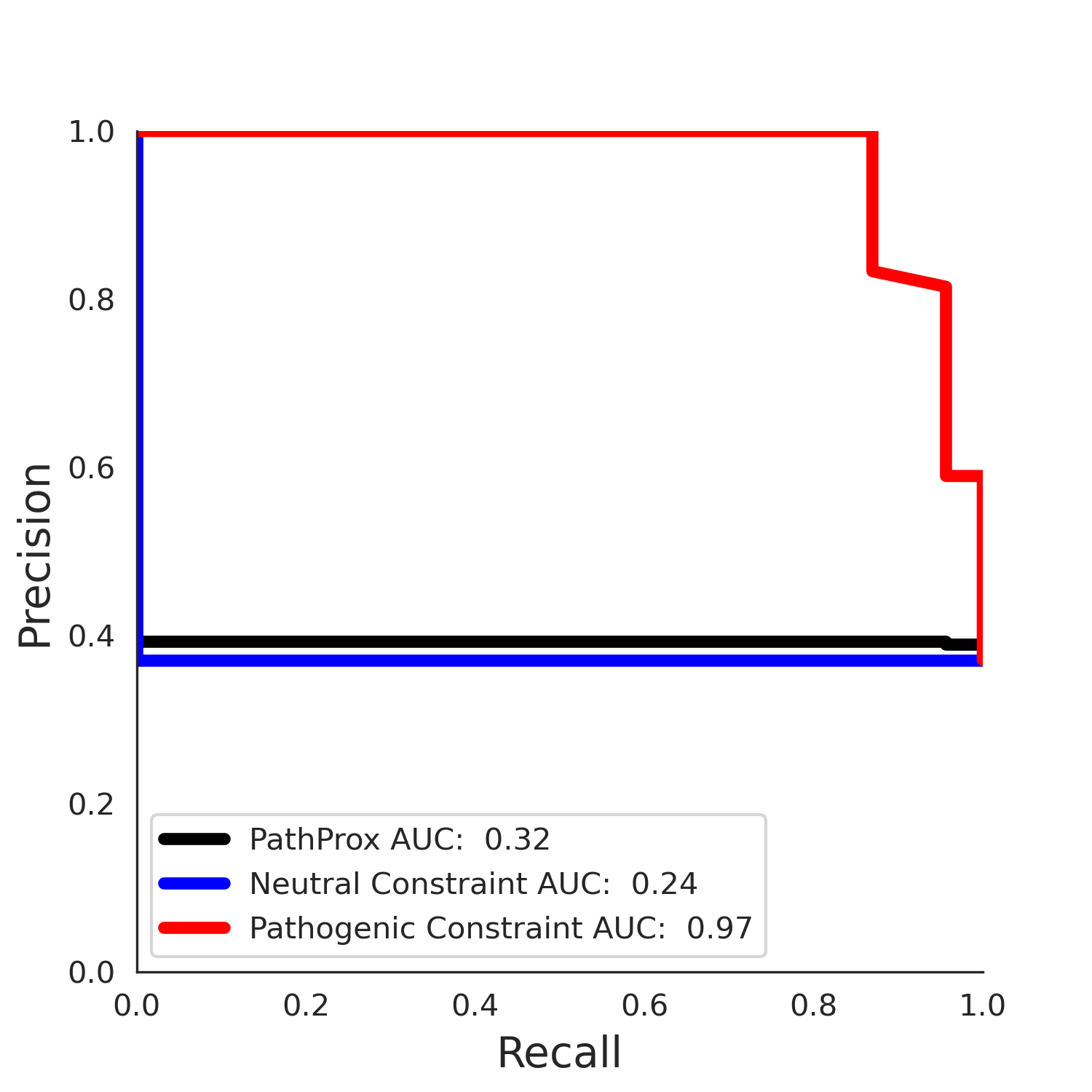
| -2.46 | 39.00 | 2.56 | 34.00 | 0.97 | 0.97 | 0.46 | 0.85 | 0.30 |
COSMIC Results - P22557-2_106_541_6hrh.1.A.B
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
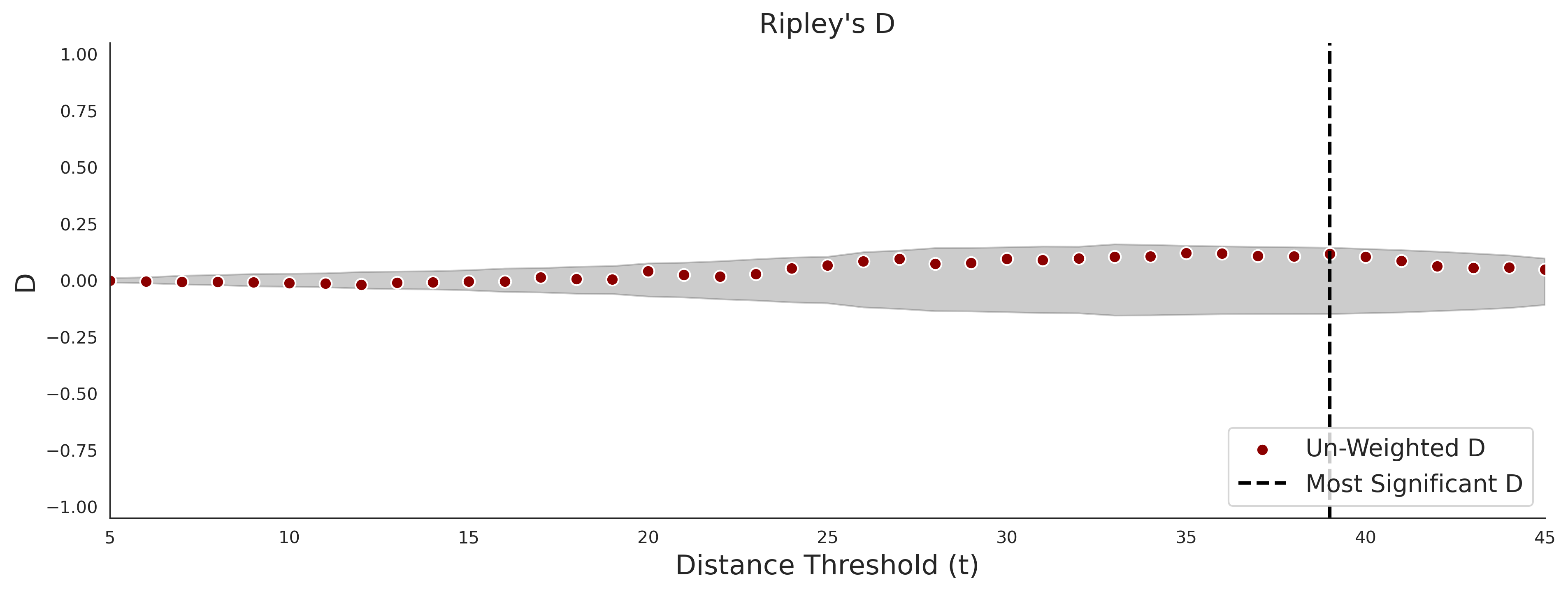
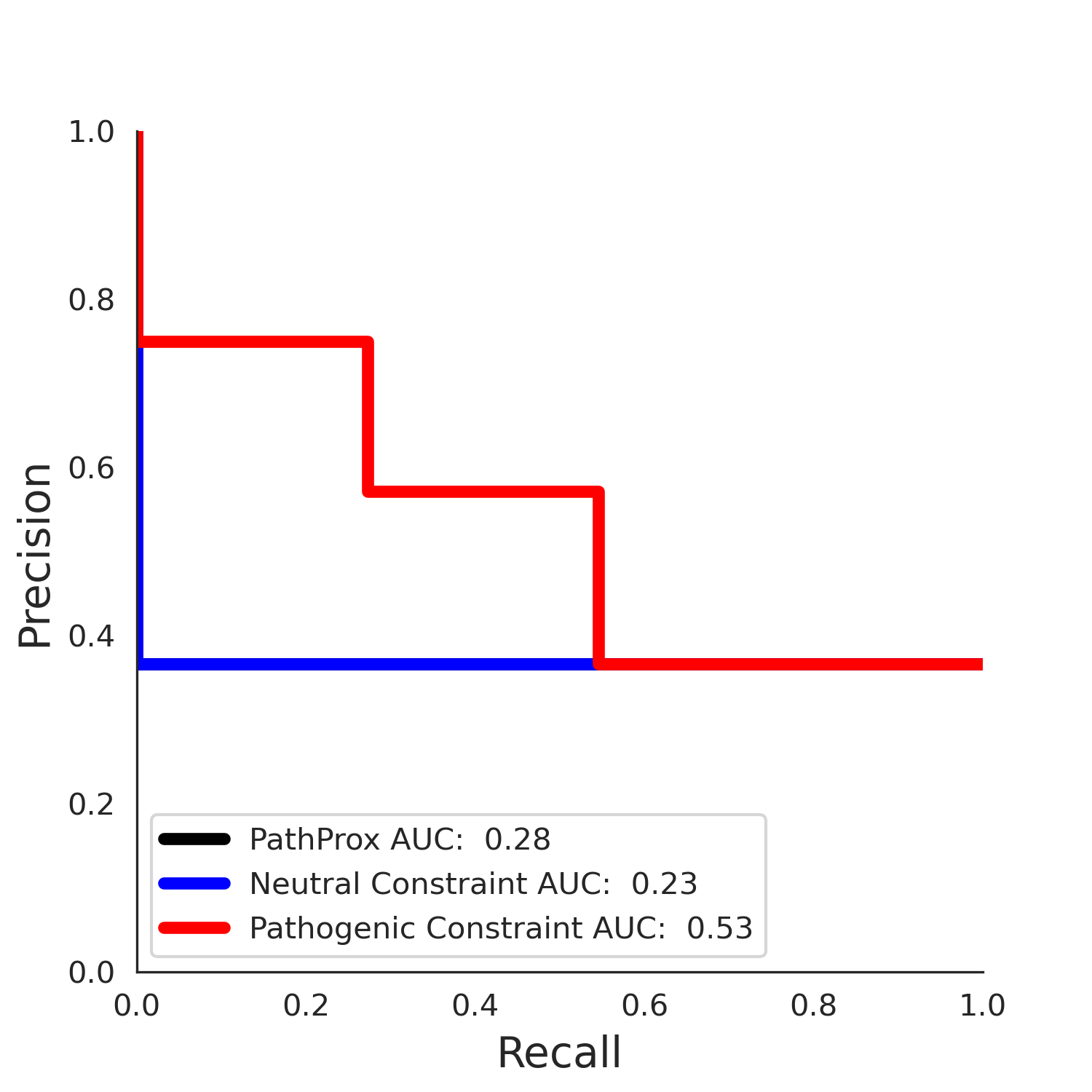
| -2.47 | 36.00 | -1.06 | 11.00 | 0.55 | 0.53 | 0.03 | 1.00 | 0.02 |
Location of A245T in P22557-2_106_541_6hrh.1.A.B (NGL Viewer)
Variant Distributions
38 gnomAD38
23 clinvar
22 COSMIC
Ripley's Univariate K
gnomAD38

clinvar
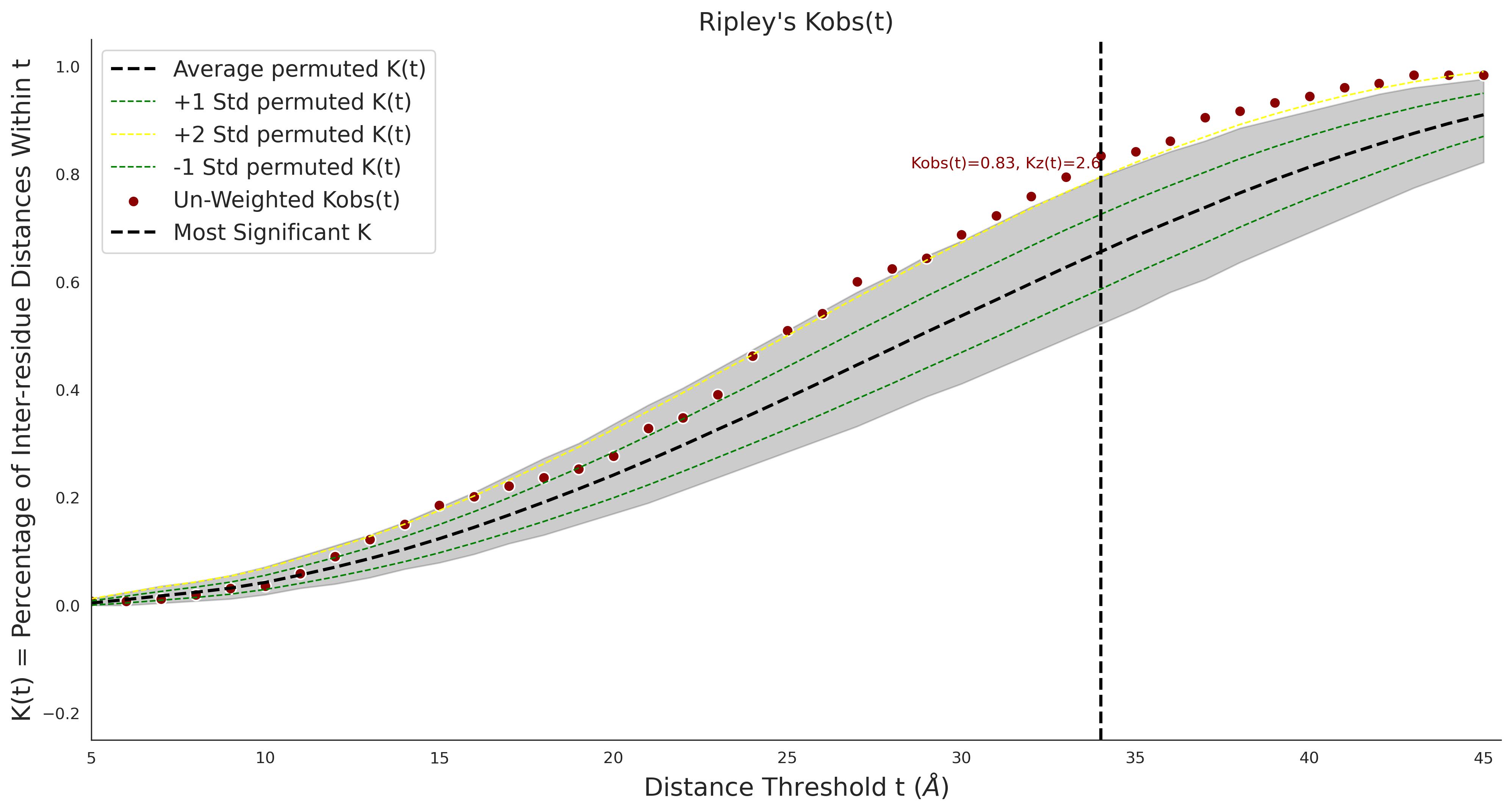
COSMIC
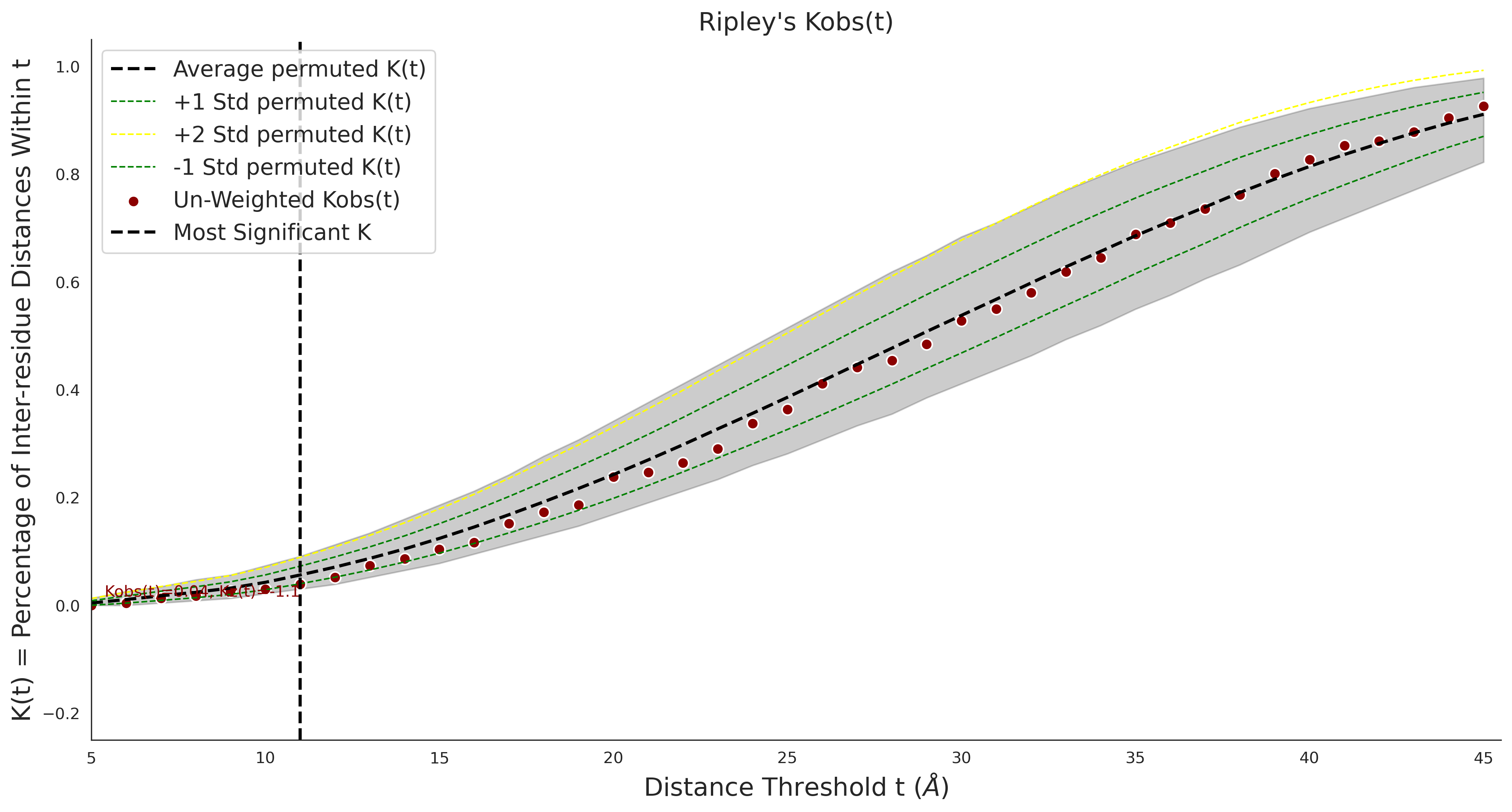
Ripley's Bivariate D
clinvar - gnomAD38
COSMIC - gnomAD38
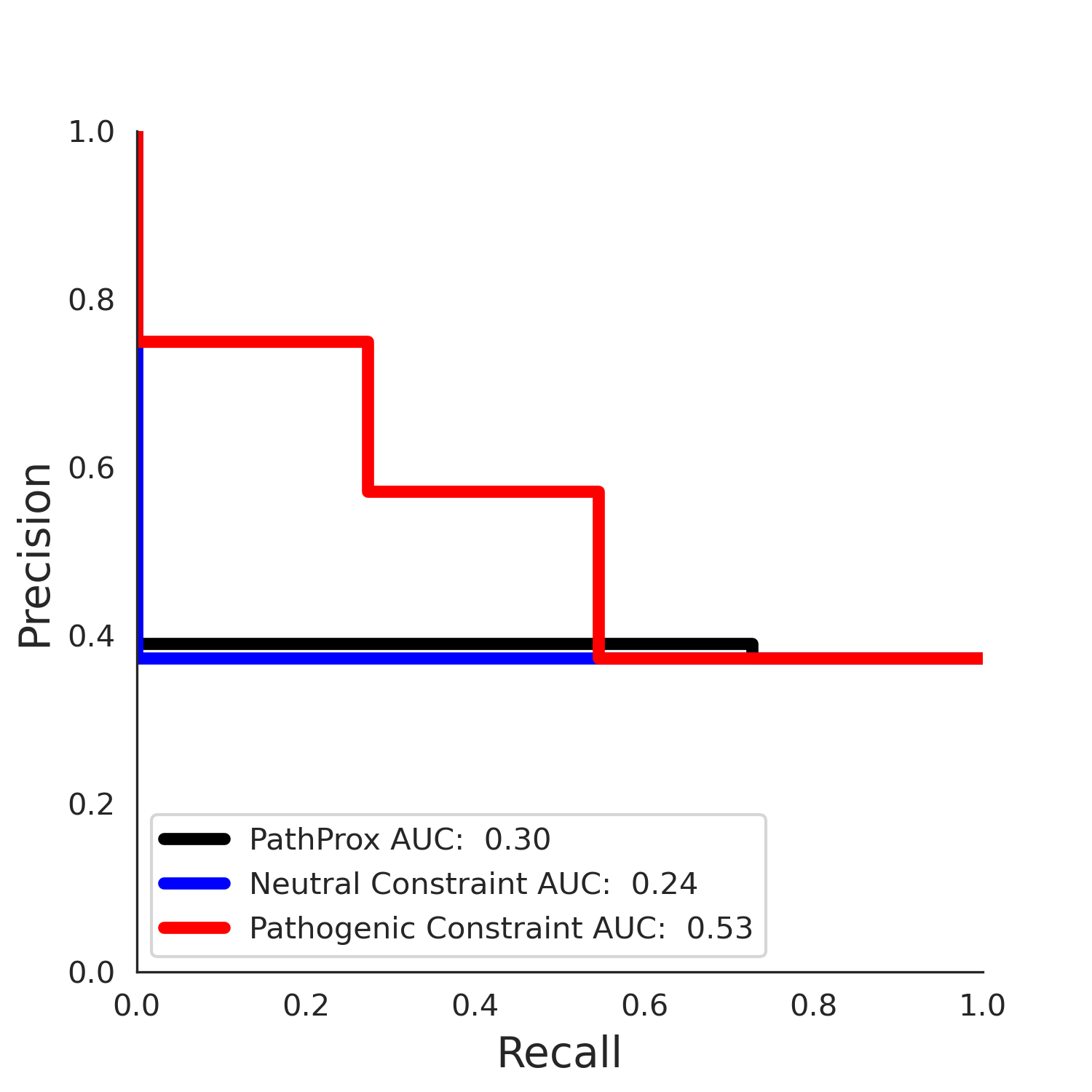
PathProx (clinvar - gnomAD38) Results
PathProx
No graphic was createdPathProx clinvar ROC
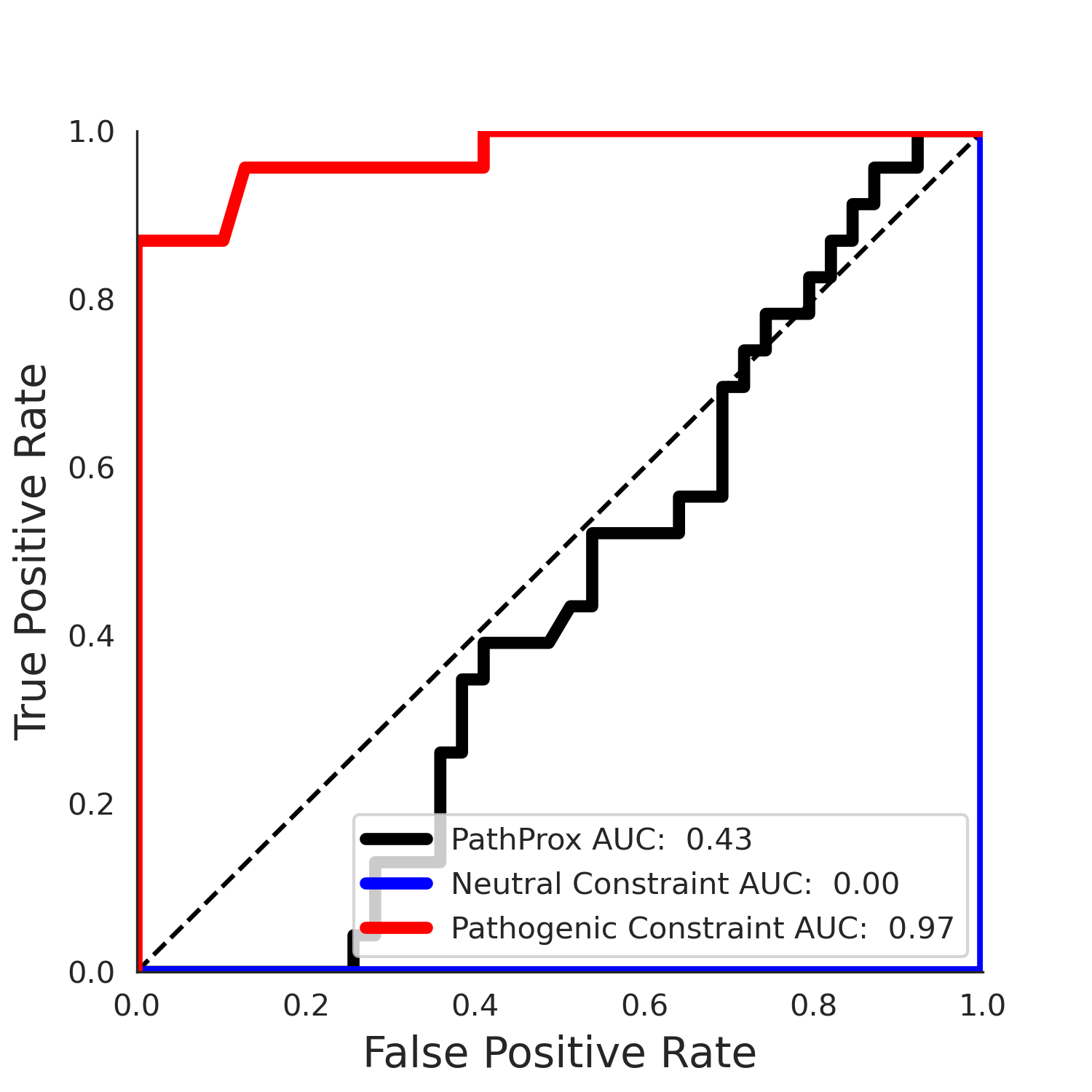
PathProx clinvar PR
PathProx (COSMIC - gnomAD38) Results
PathProx
No graphic was createdPathProx COSMIC ROC
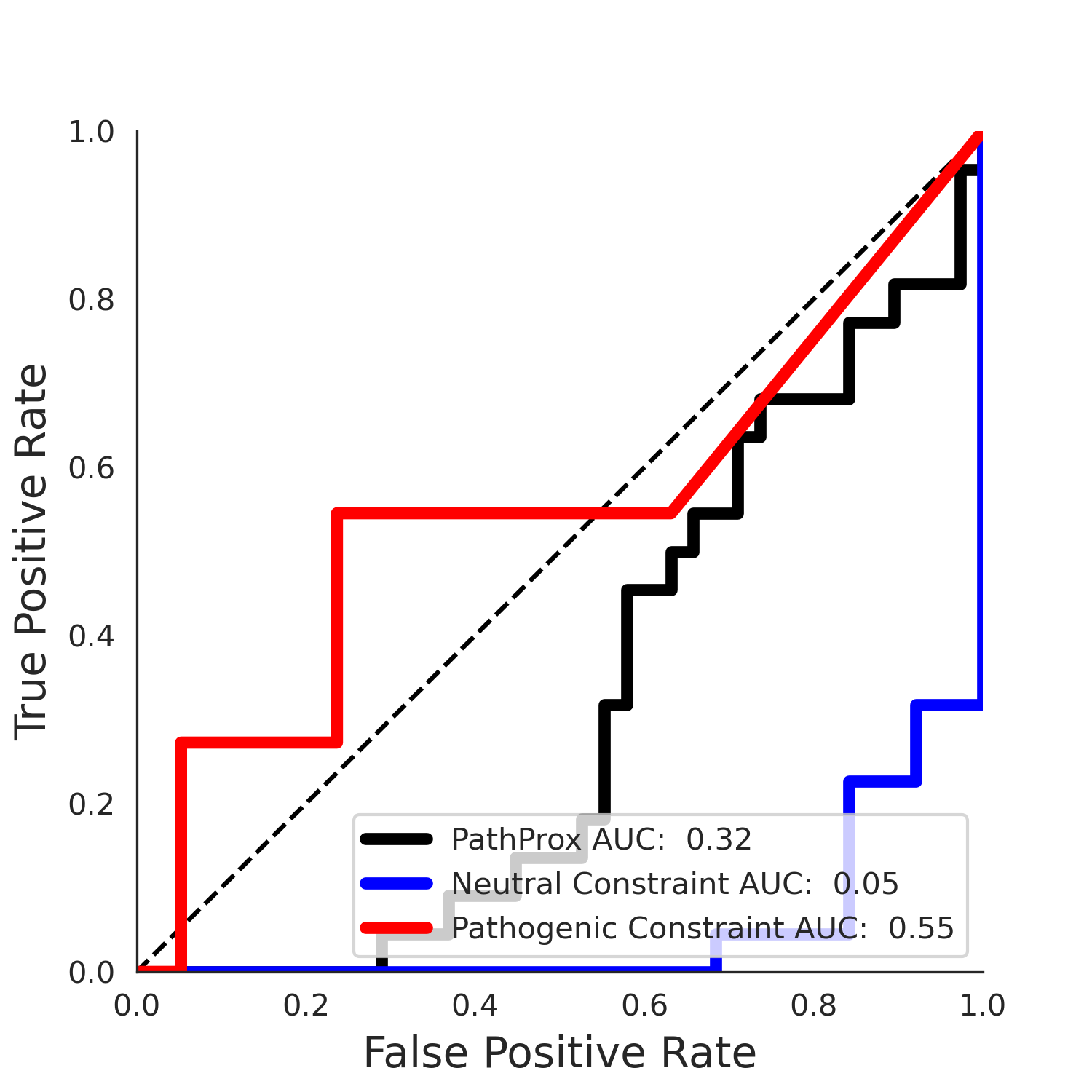
PathProx COSMIC PR
P22557-2_106_541_6hrh.1.A.B trimer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| swiss | P22557-2_106_541_6hrh.1.A | B | 6hrh.1.A | 99.78 | 1.0 | 106 | 541 | 0.06 | 0.797 | 0.02 | 0.494 |
clinvar Results - P22557-2_106_541_6hrh.1.A.B
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -3.42 | 32.00 | 2.38 | 15.00 | 0.85 | 0.80 | 0.06 | 0.99 | 0.03 |
COSMIC Results - P22557-2_106_541_6hrh.1.A.B
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -3.10 | 32.00 | -1.50 | 11.00 | 0.54 | 0.49 | 0.02 | 1.00 | 0.01 |
Location of A245T in P22557-2_106_541_6hrh.1.A.B (NGL Viewer)
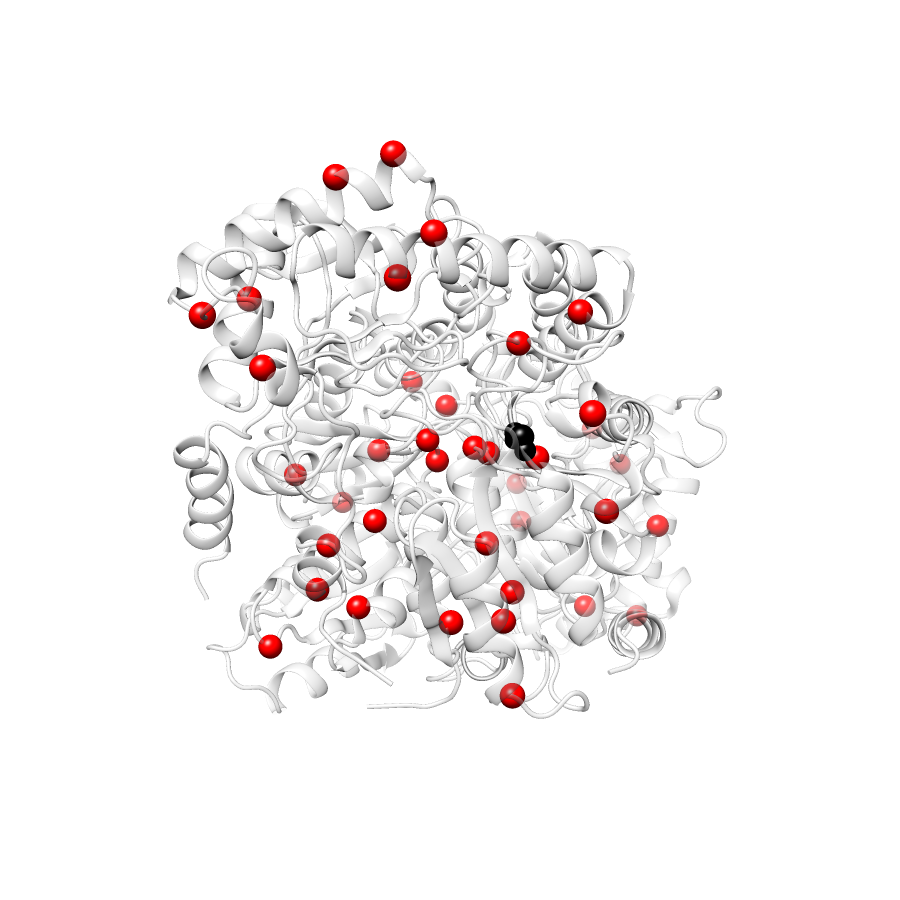
Variant Distributions
76 gnomAD38

46 clinvar
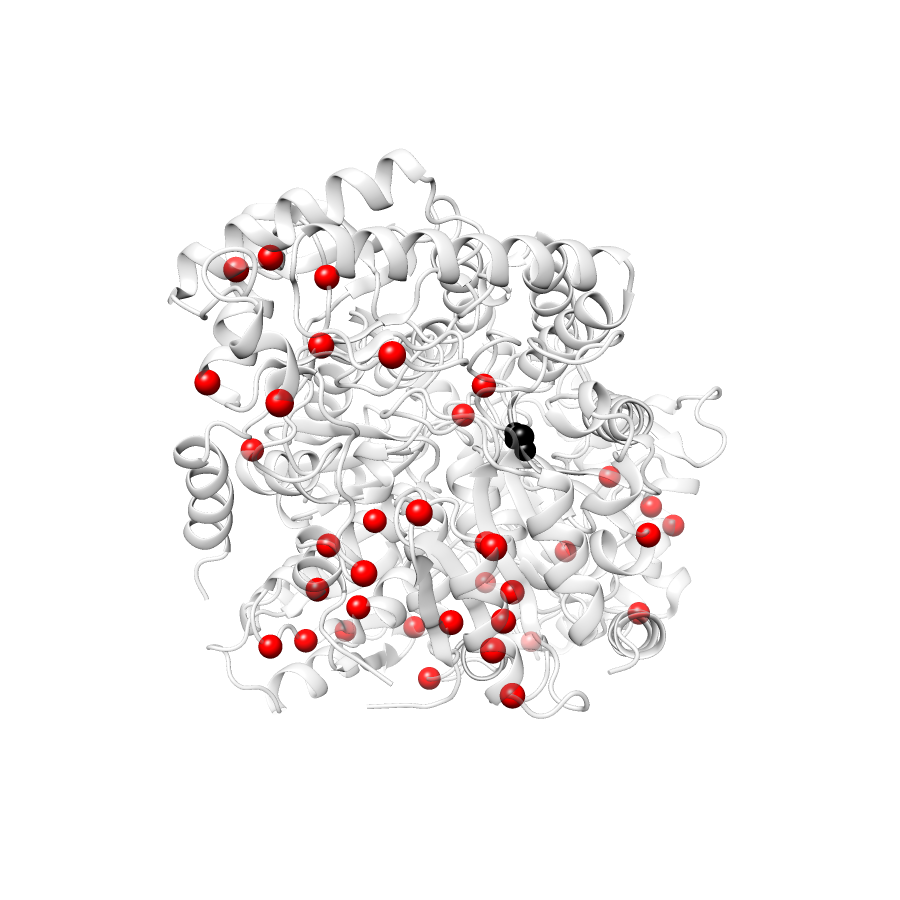
44 COSMIC
Ripley's Univariate K
gnomAD38
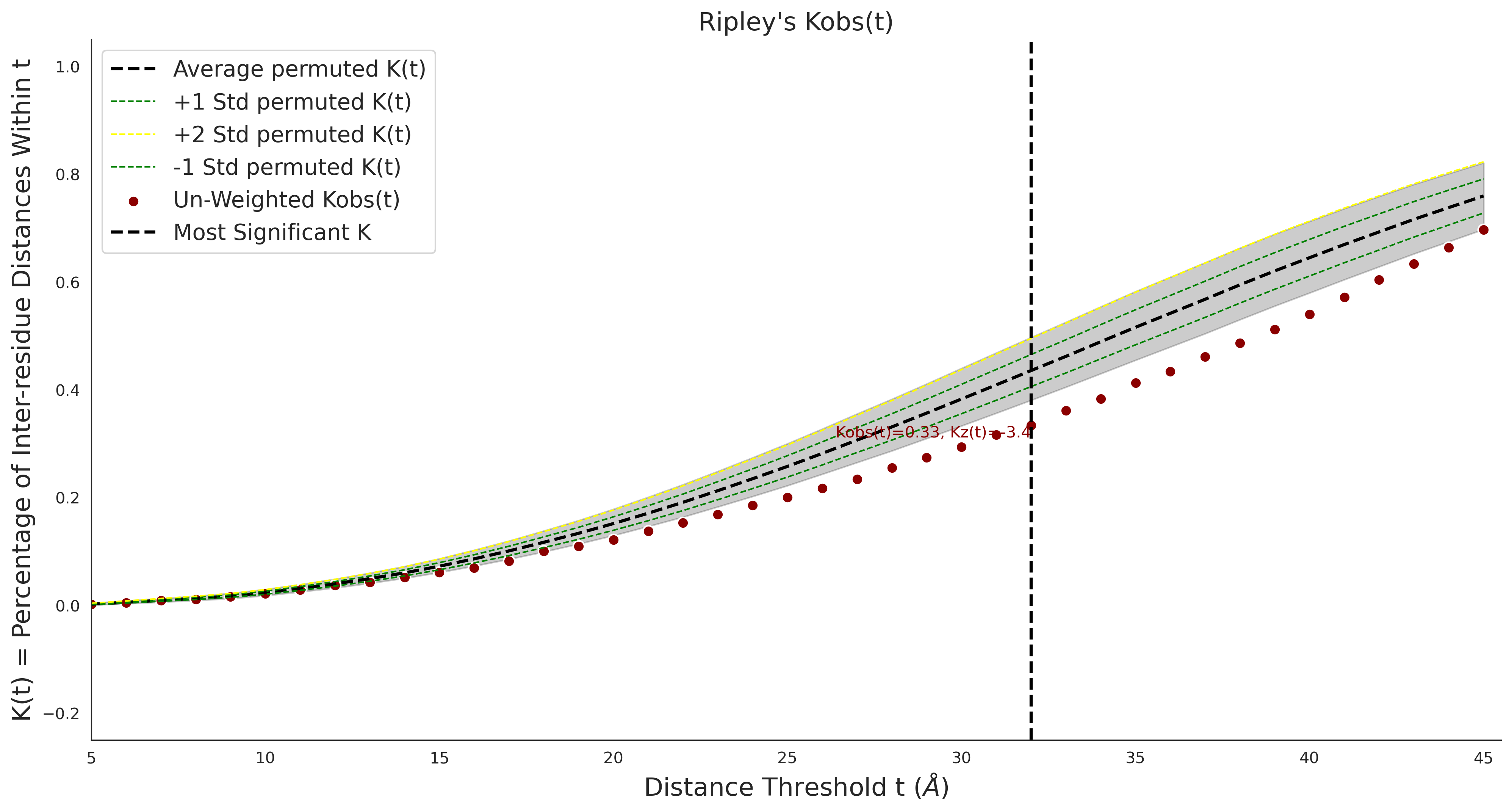
clinvar
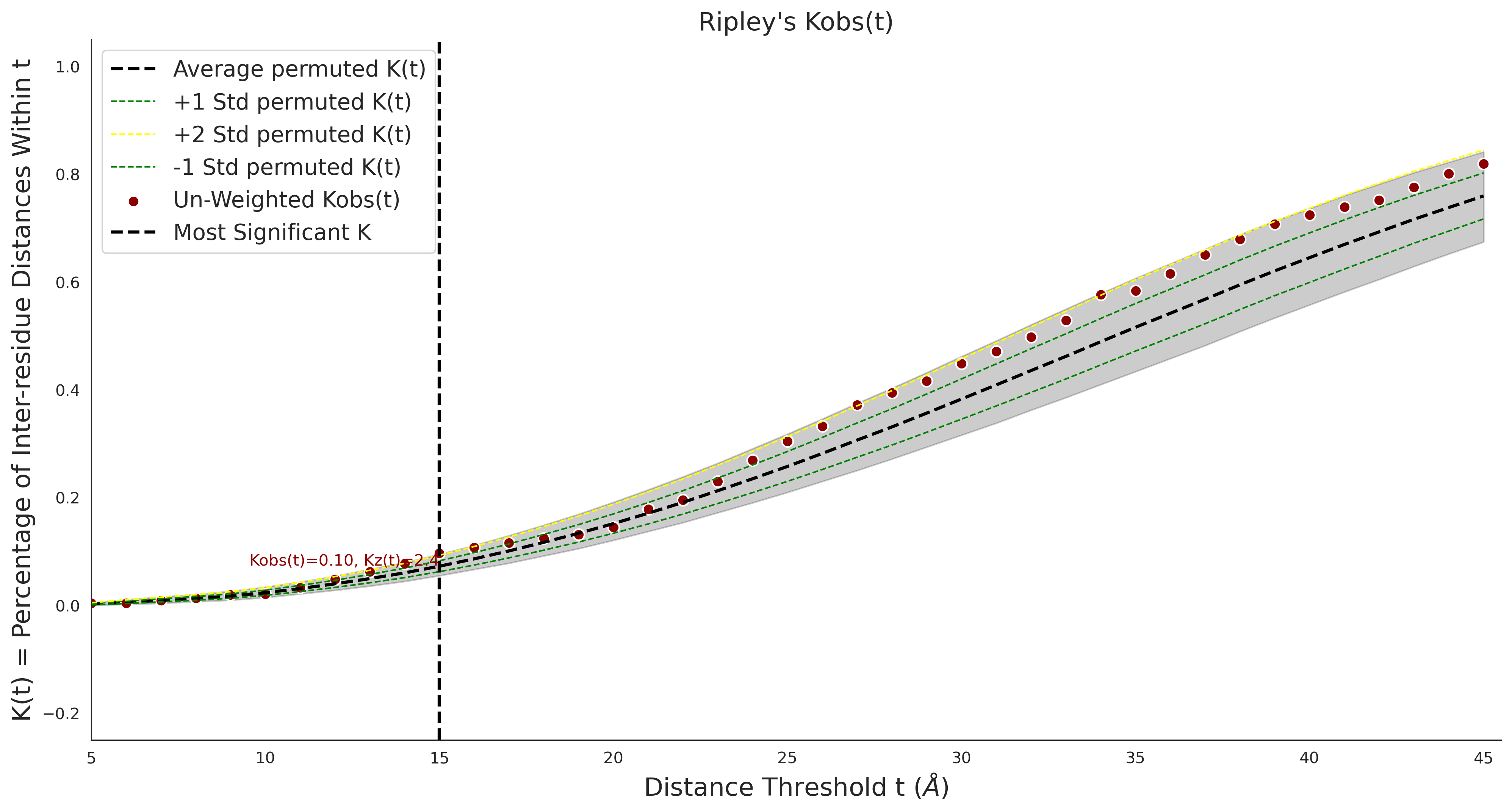
COSMIC
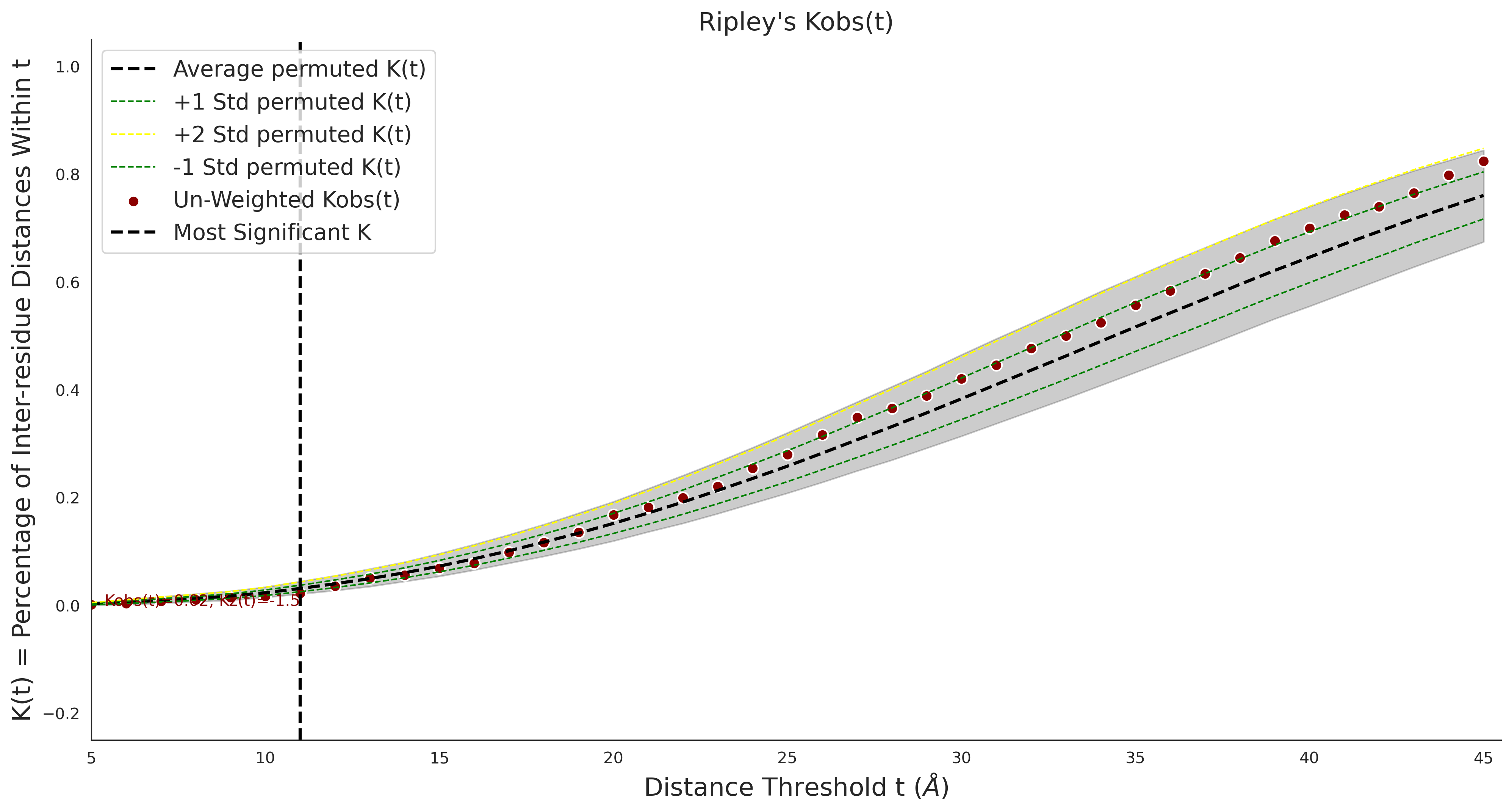
Ripley's Bivariate D
clinvar - gnomAD38
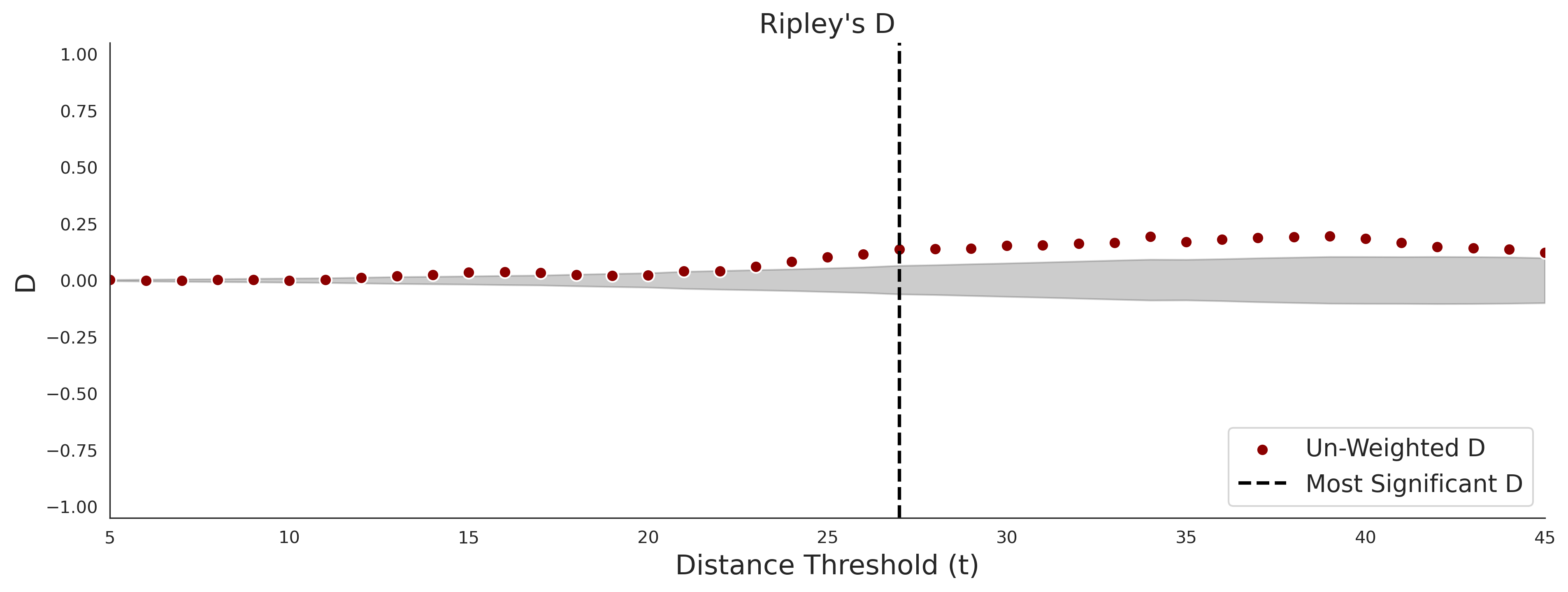
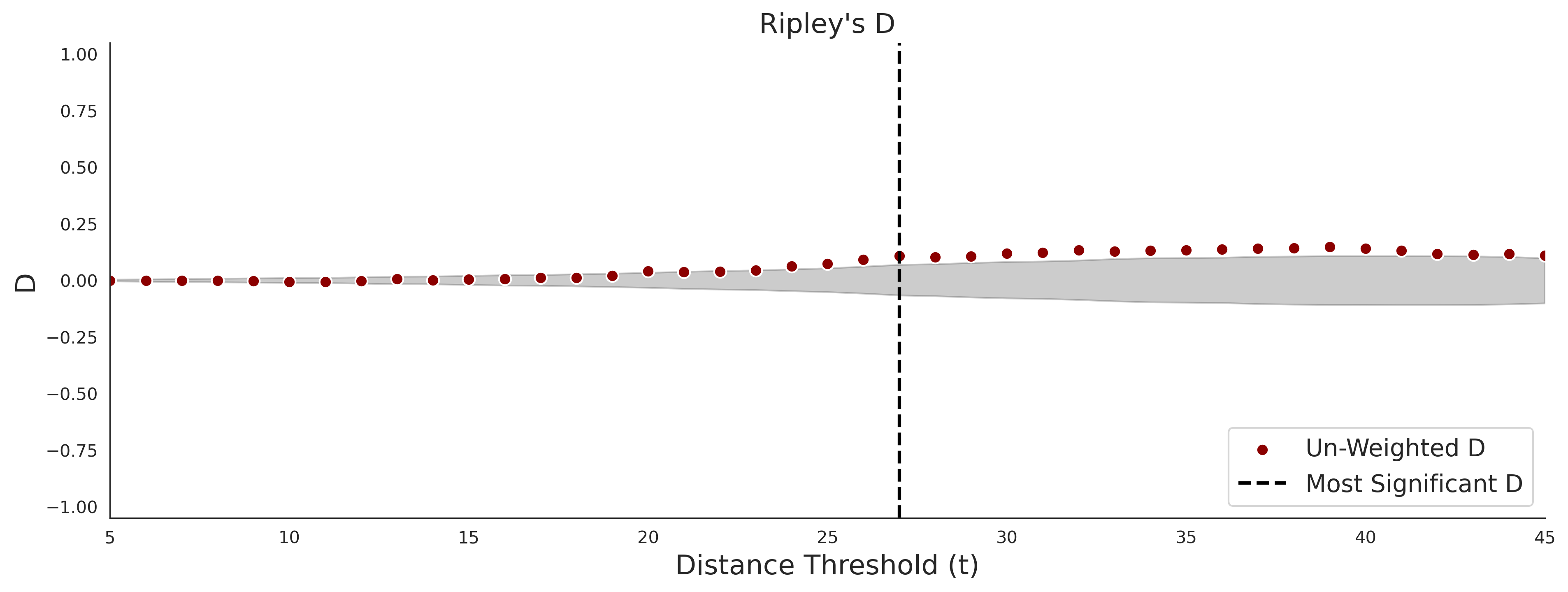
COSMIC - gnomAD38
PathProx (clinvar - gnomAD38) Results
PathProx

PathProx clinvar ROC
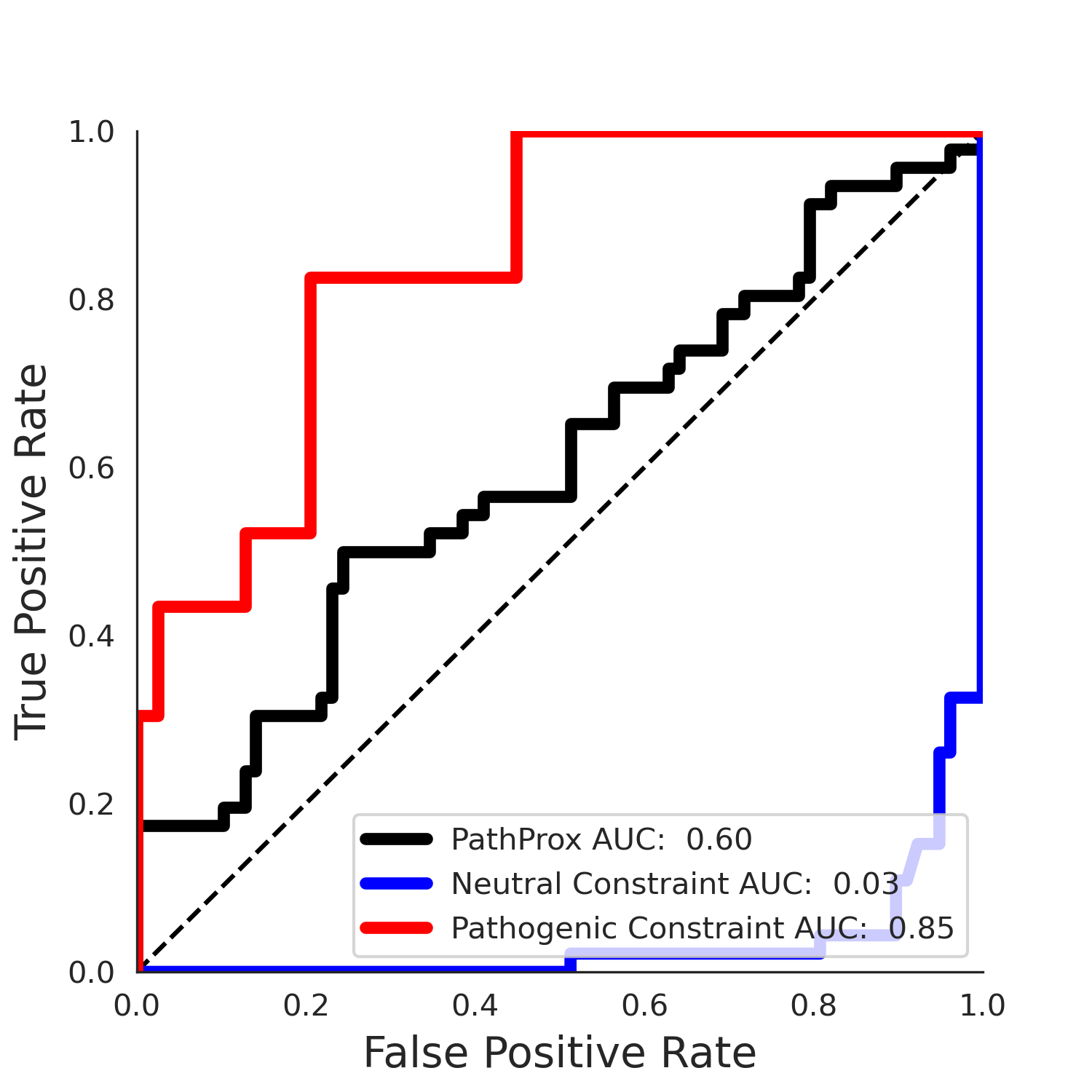
PathProx clinvar PR
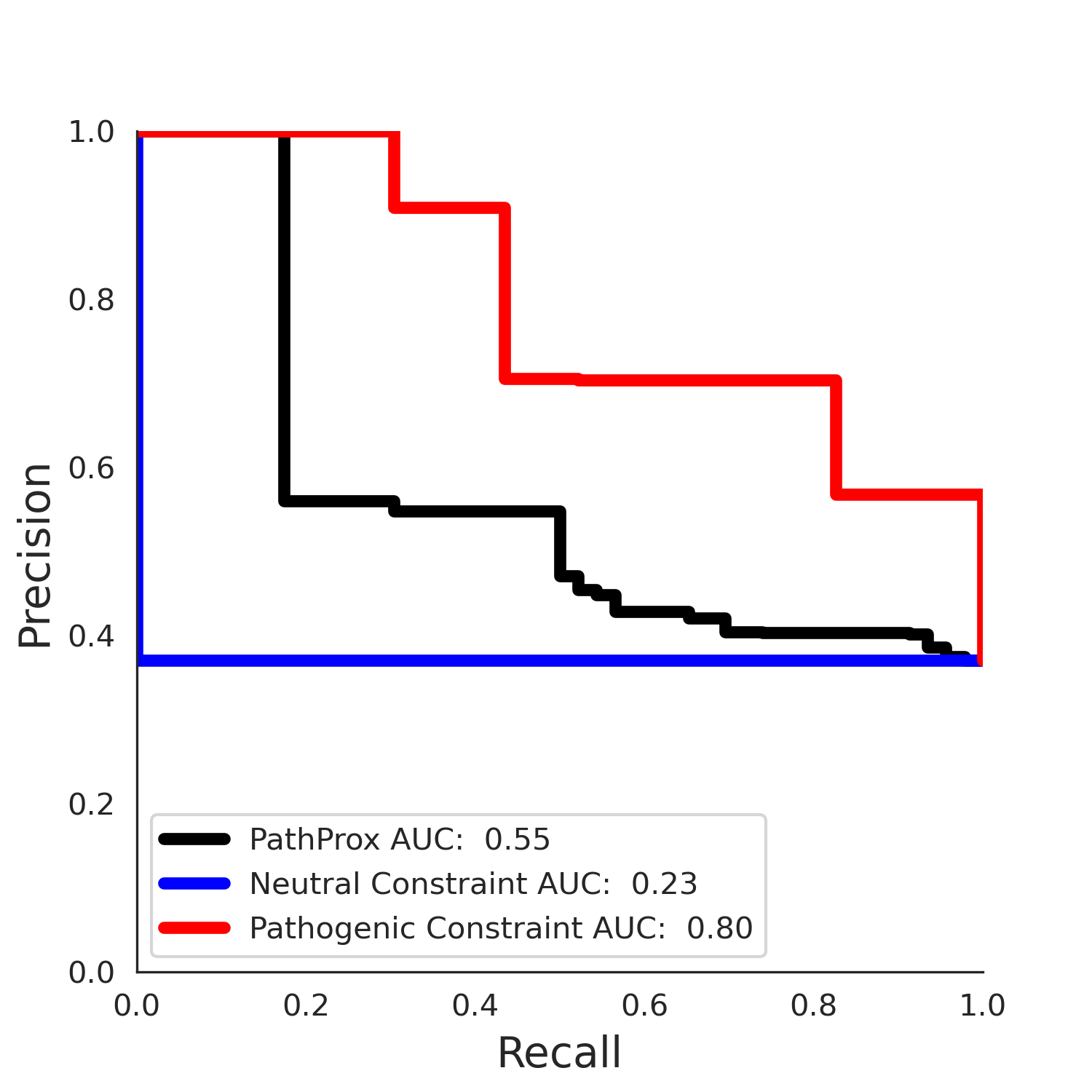
PathProx (COSMIC - gnomAD38) Results
PathProx

PathProx COSMIC ROC
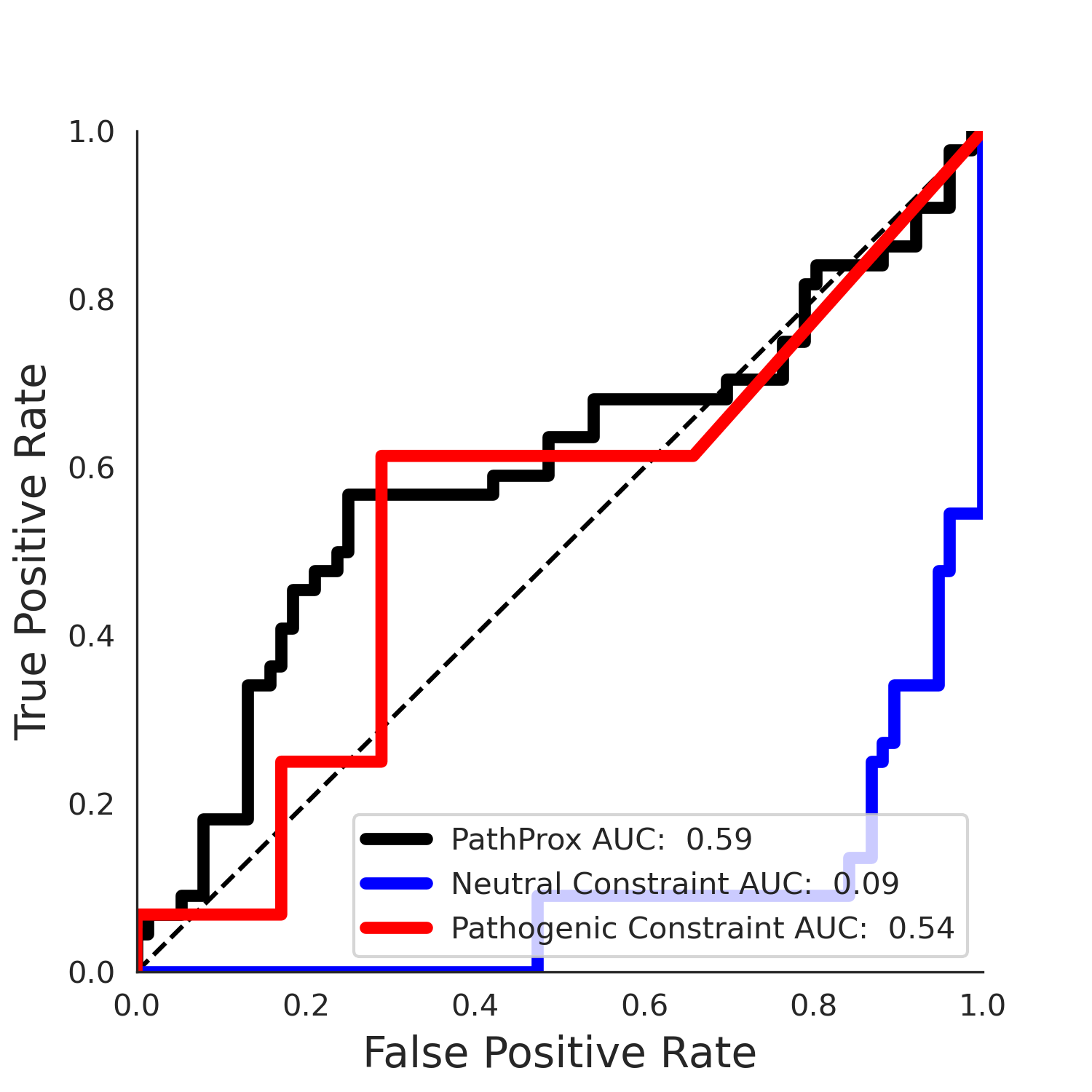
PathProx COSMIC PR
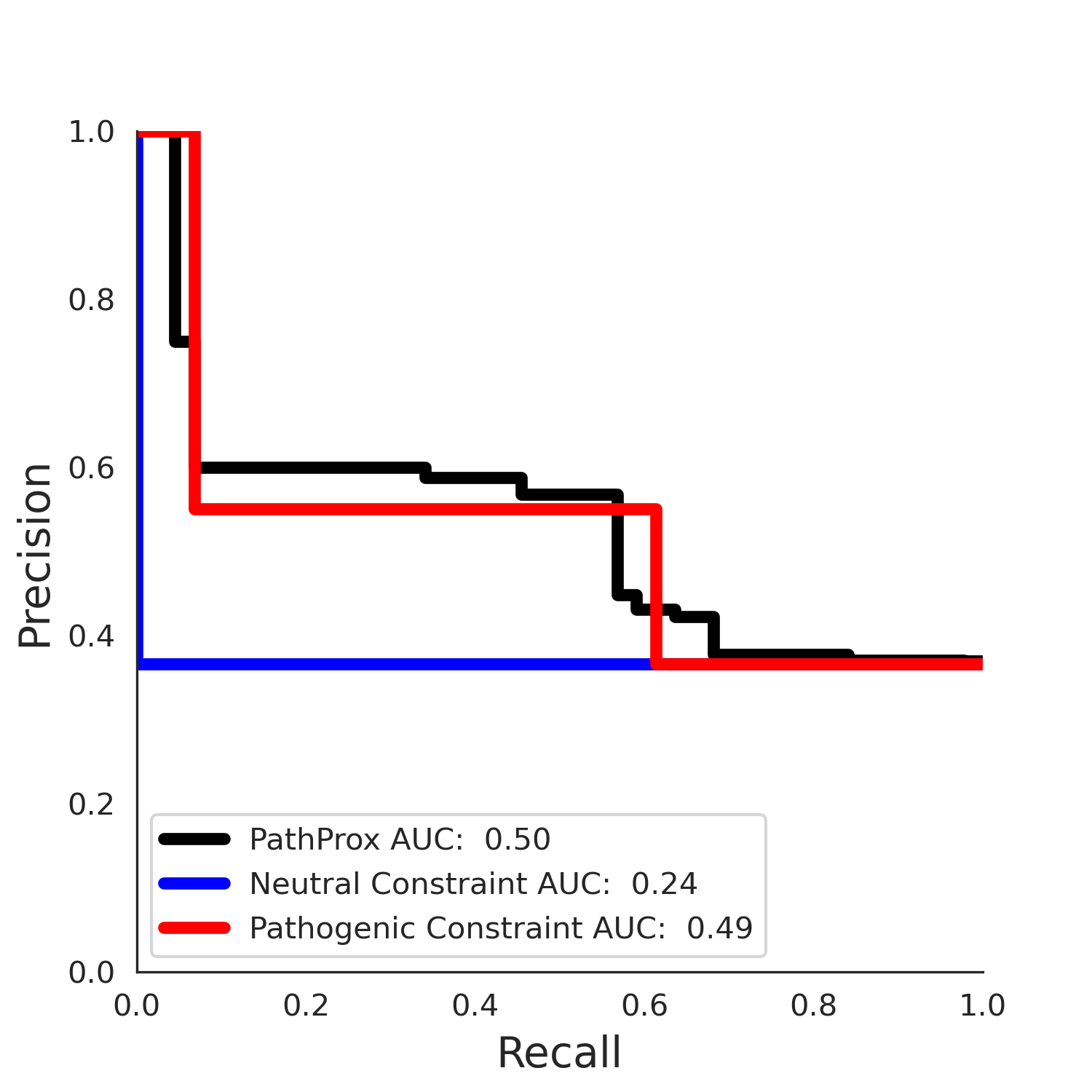
ENSP00000337131.4_1.A monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| modbase | ENSP00000337131.4_1 | A | 5qr1 | 99.0 | 1.0 | 103 | 541 | 17.06 | 11.32 | 0.5 | 0.958 | -0.01 | 0.471 |
clinvar Results - ENSP00000337131.4_1.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -2.14 | 38.00 | 2.66 | 33.00 | 0.97 | 0.96 | 0.50 | 0.87 | 0.30 |
COSMIC Results - ENSP00000337131.4_1.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -2.09 | 38.00 | -1.09 | 8.00 | 0.49 | 0.47 | -0.01 | 1.00 | 0.00 |
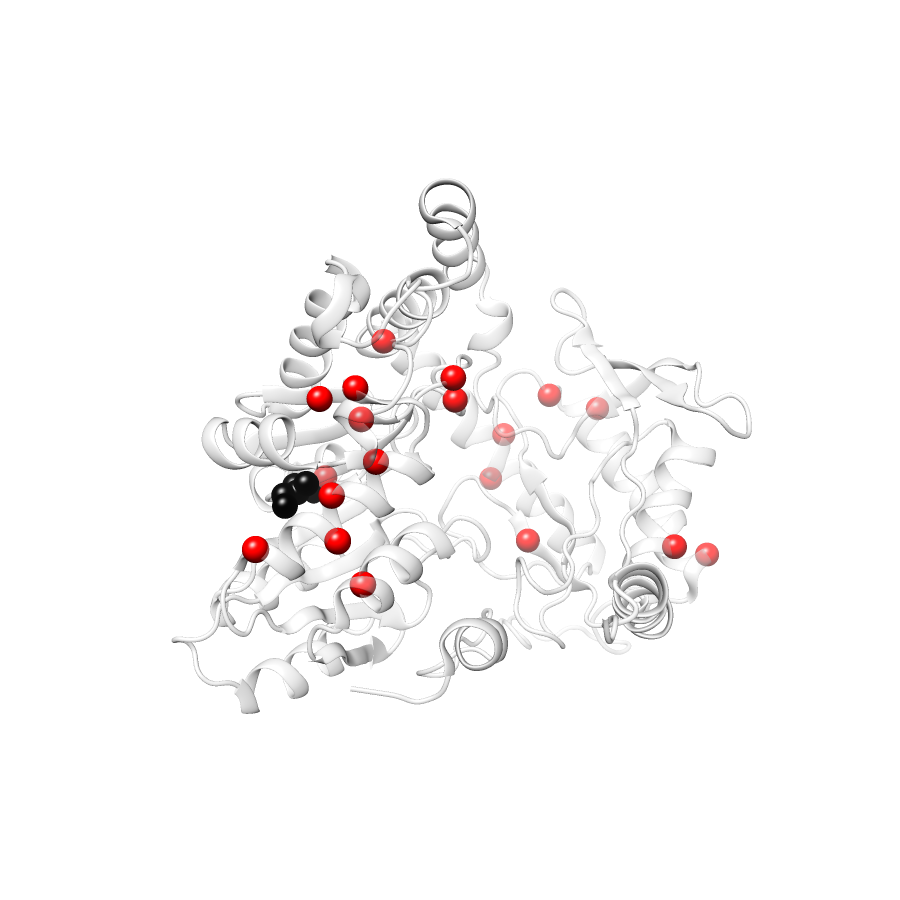
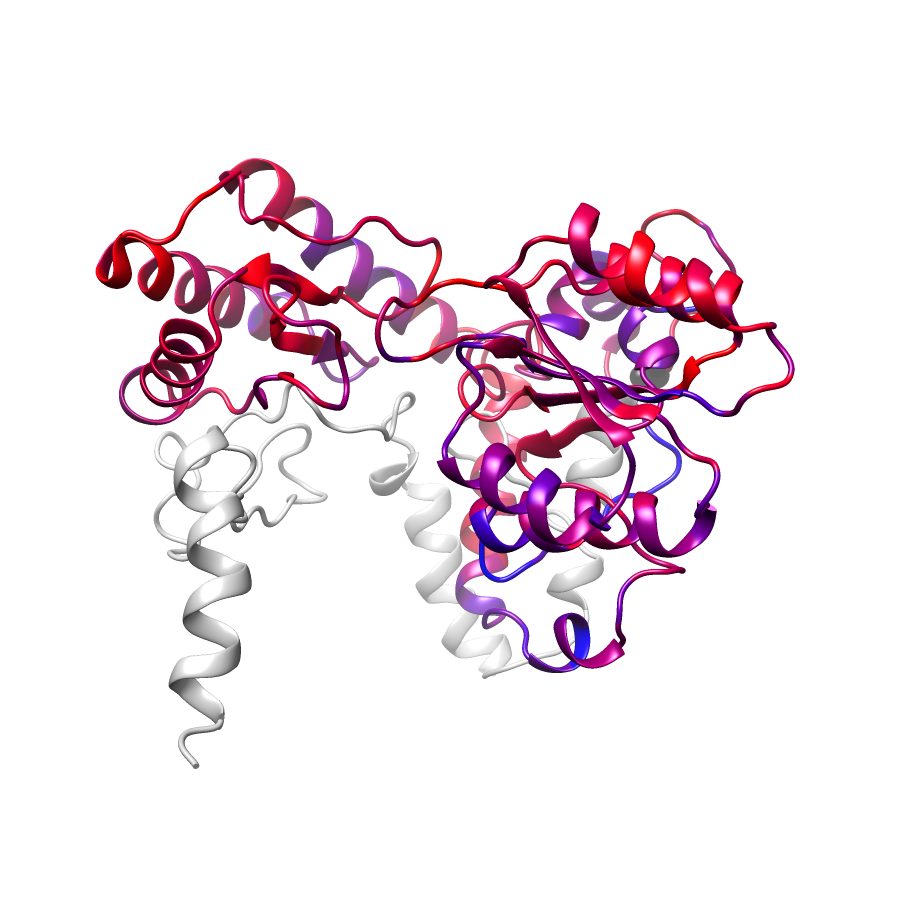
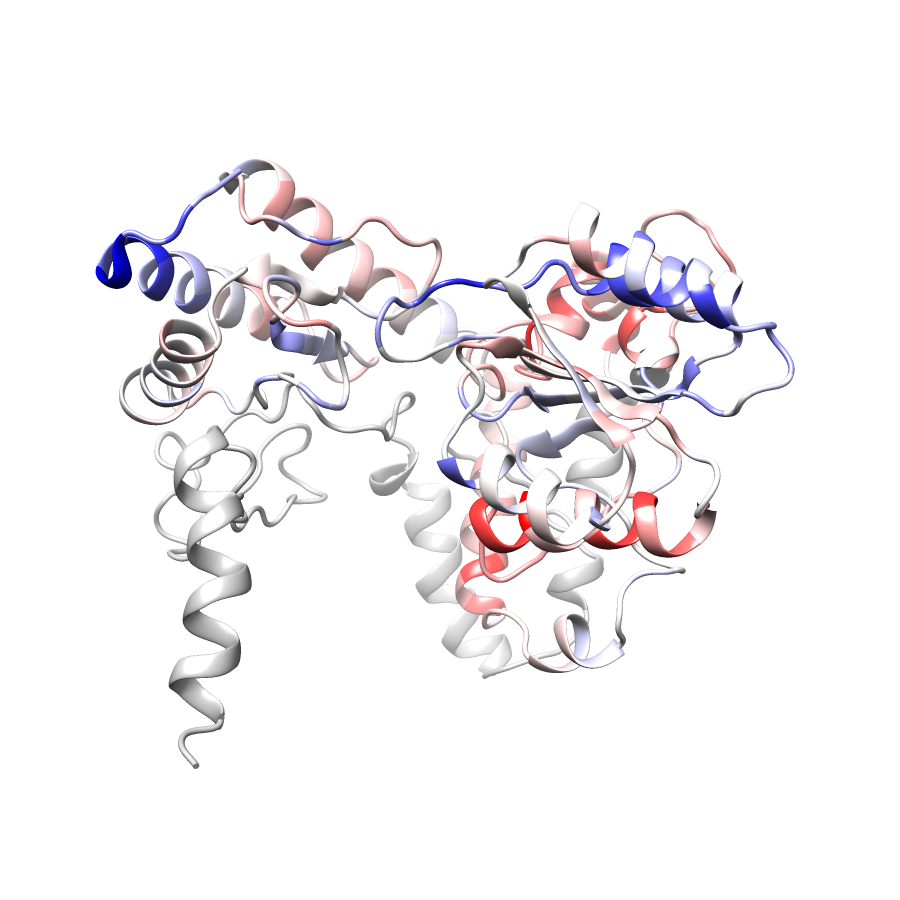
Location of A245T in ENSP00000337131.4_1.A (NGL Viewer)
Variant Distributions
38 gnomAD38
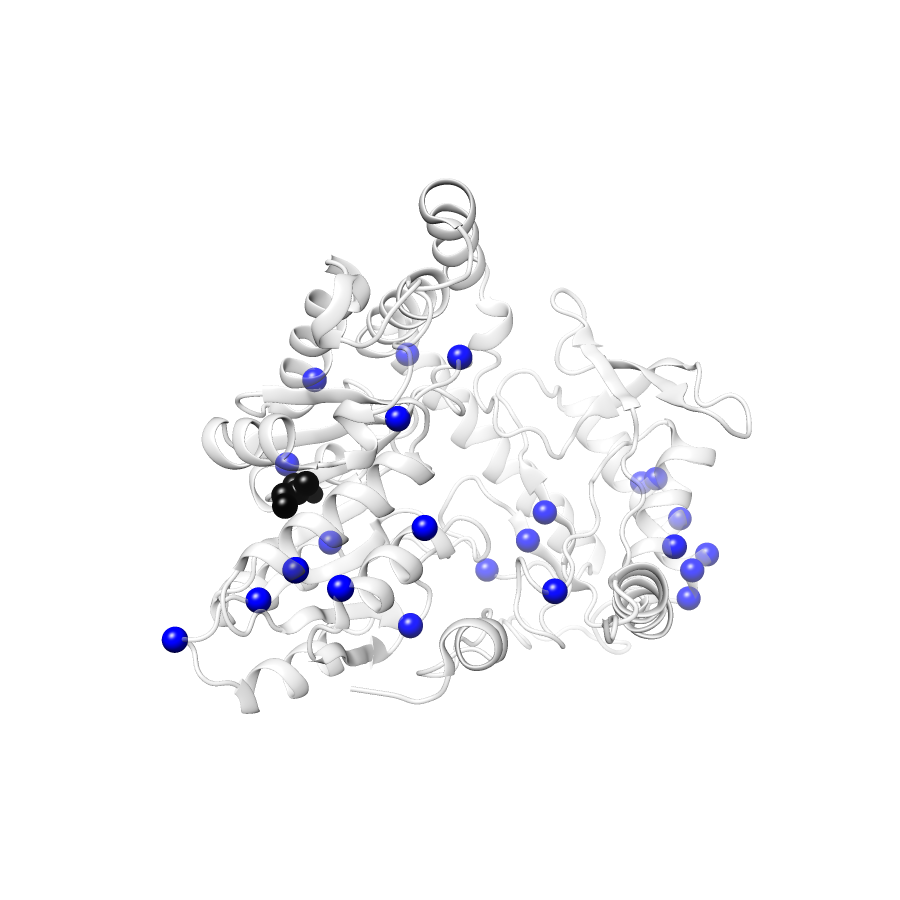
23 clinvar
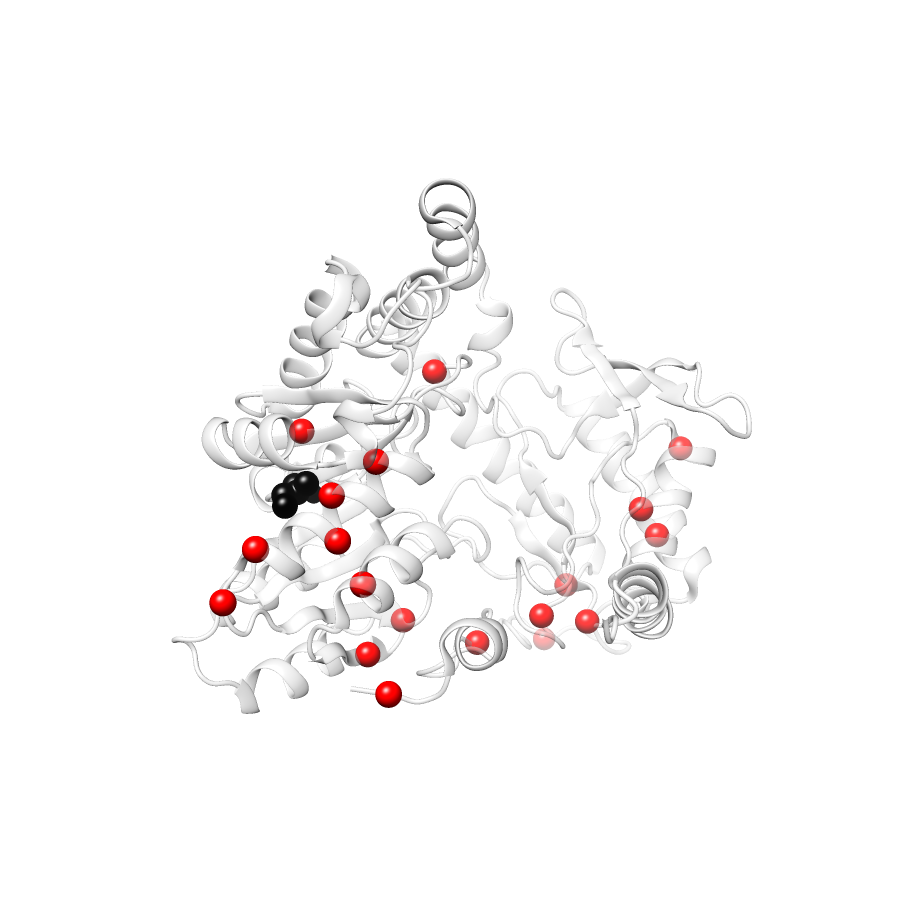
22 COSMIC
Ripley's Univariate K
gnomAD38
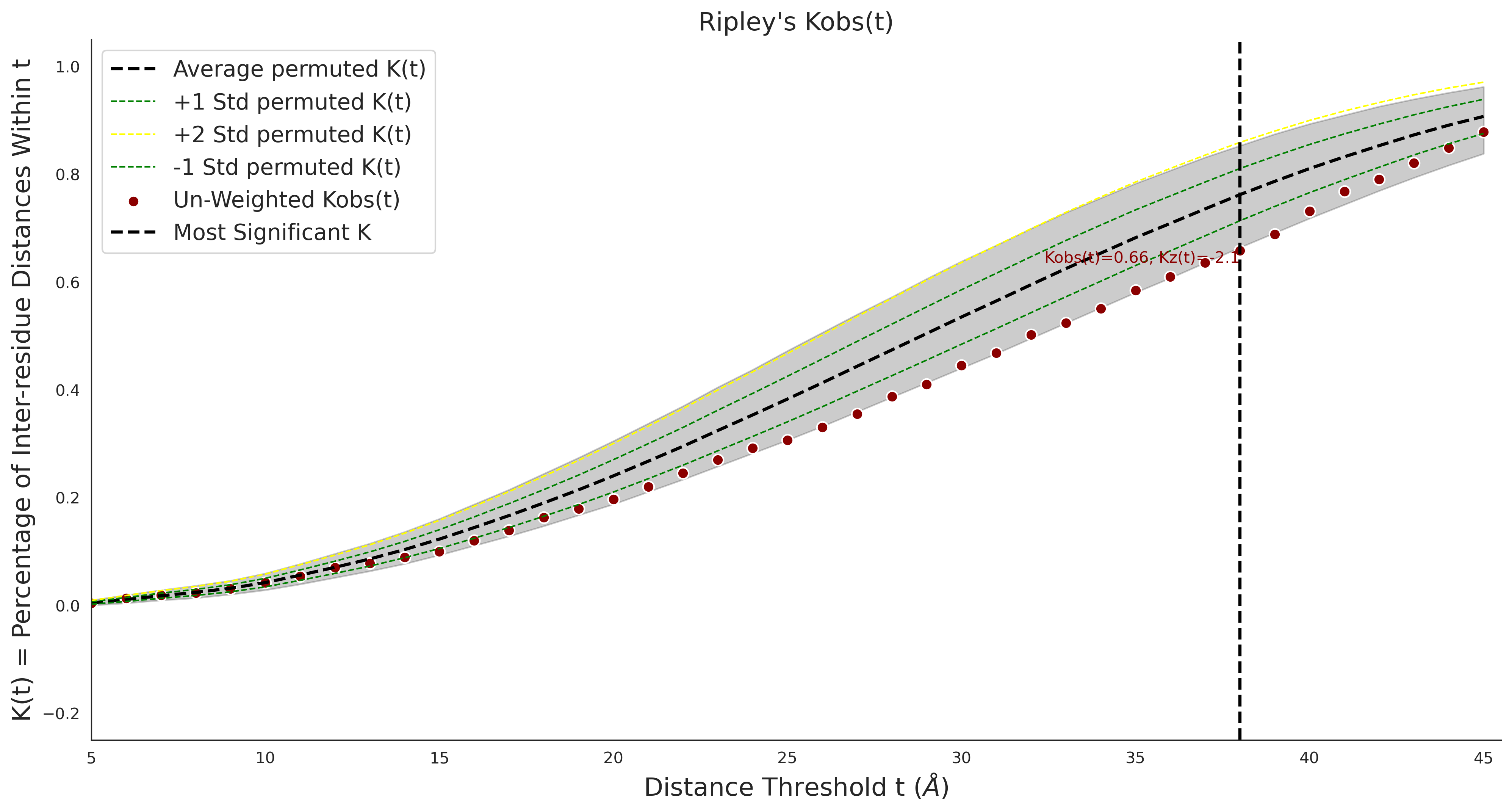
clinvar
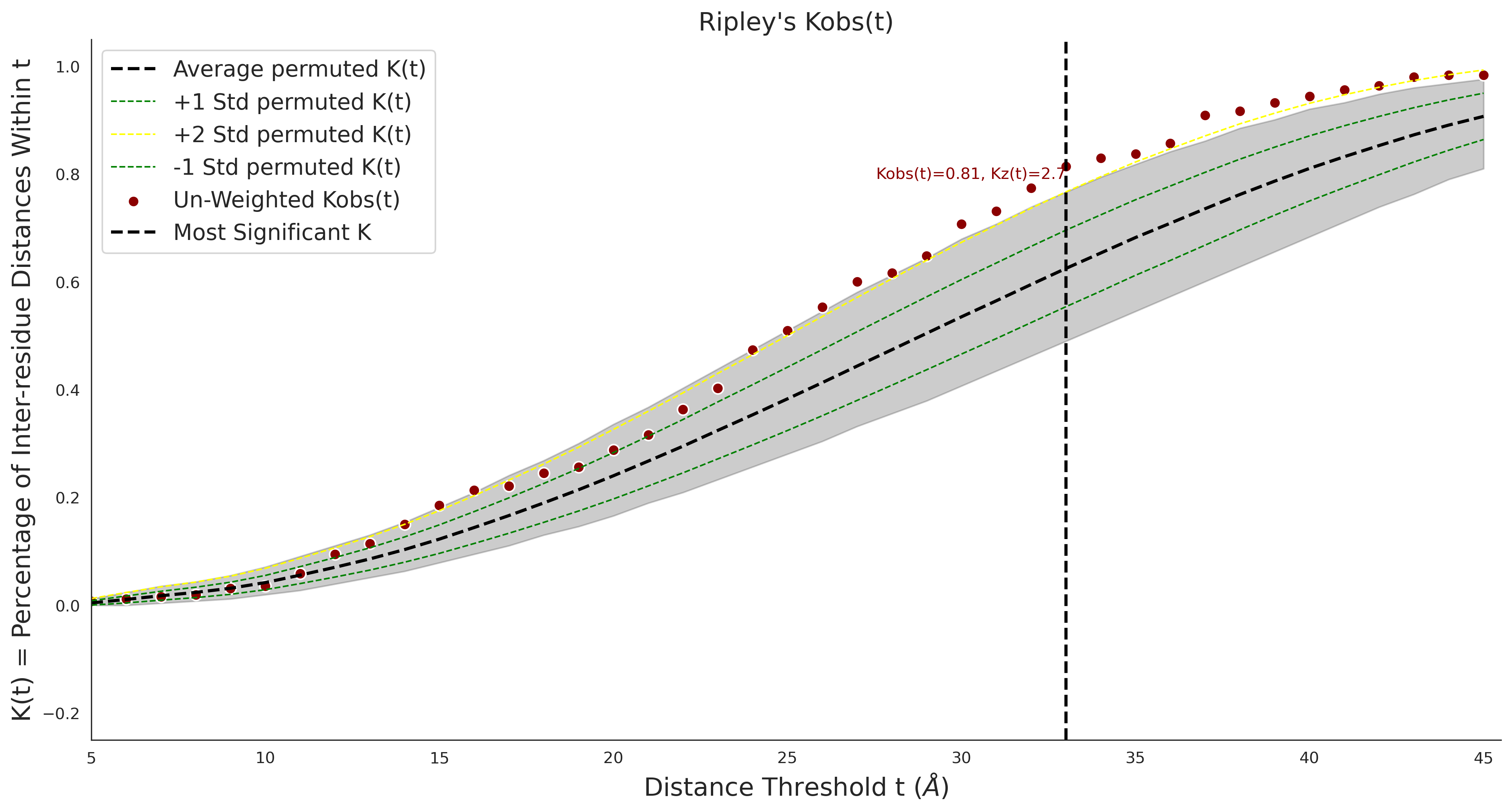
COSMIC
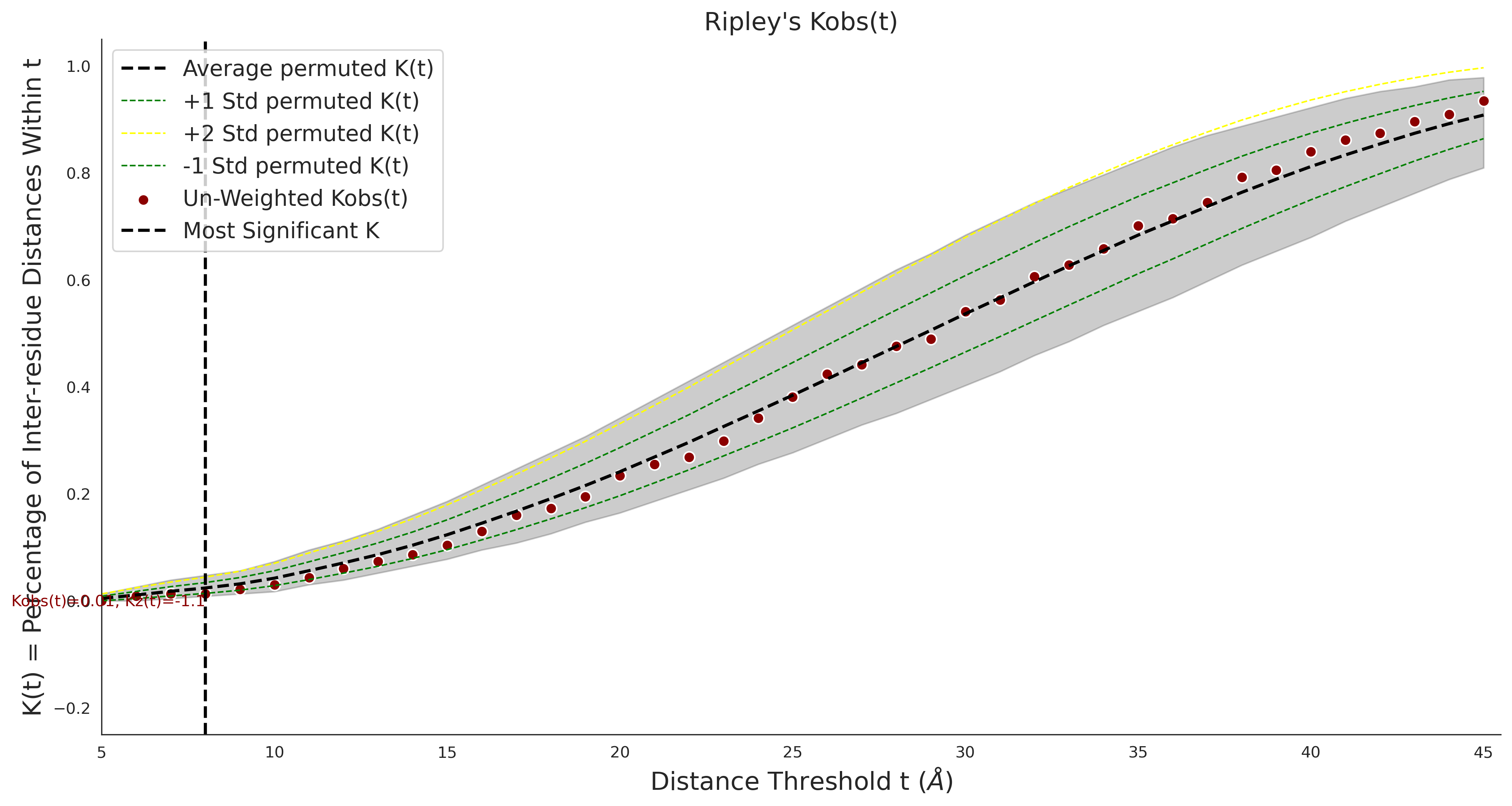
Ripley's Bivariate D
clinvar - gnomAD38
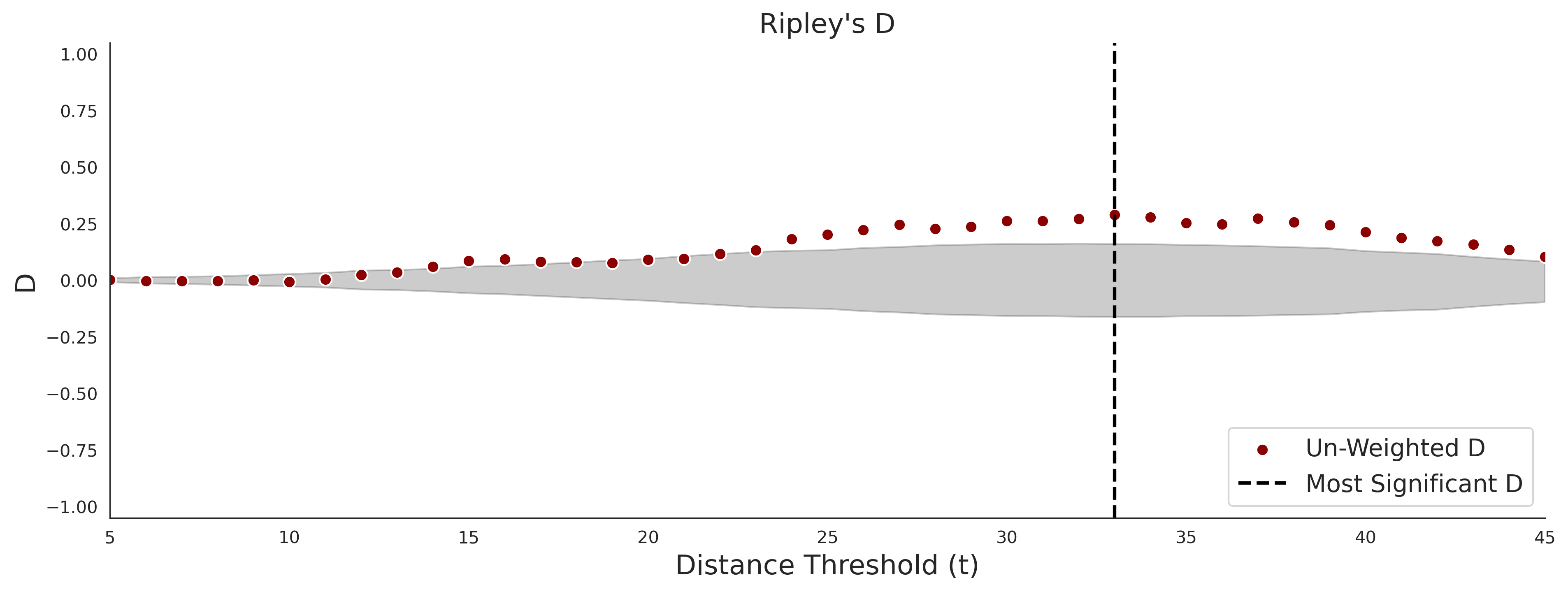
COSMIC - gnomAD38
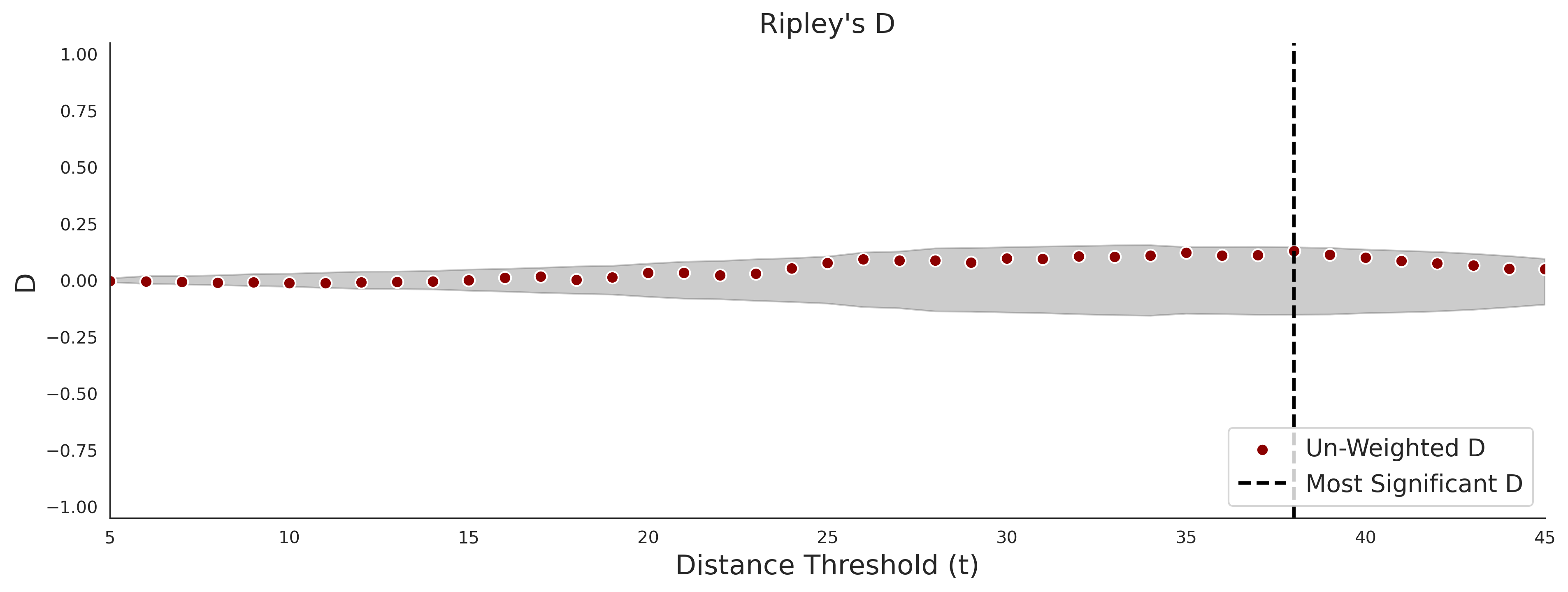
PathProx (clinvar - gnomAD38) Results
PathProx
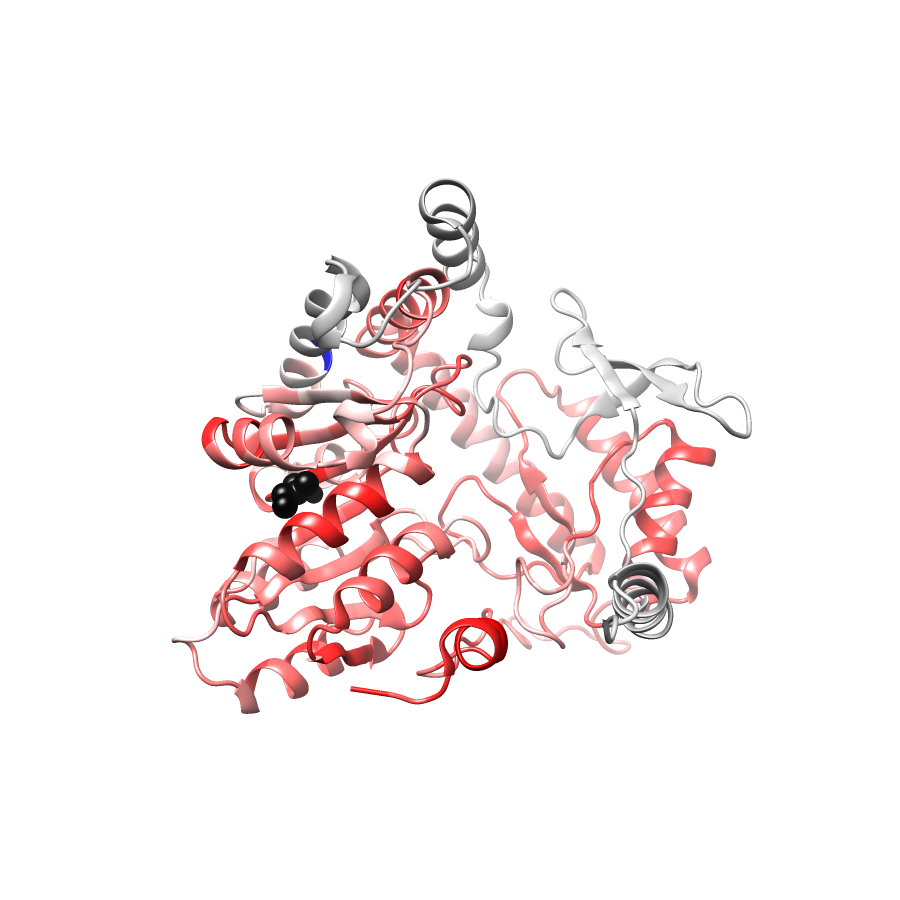
PathProx clinvar ROC
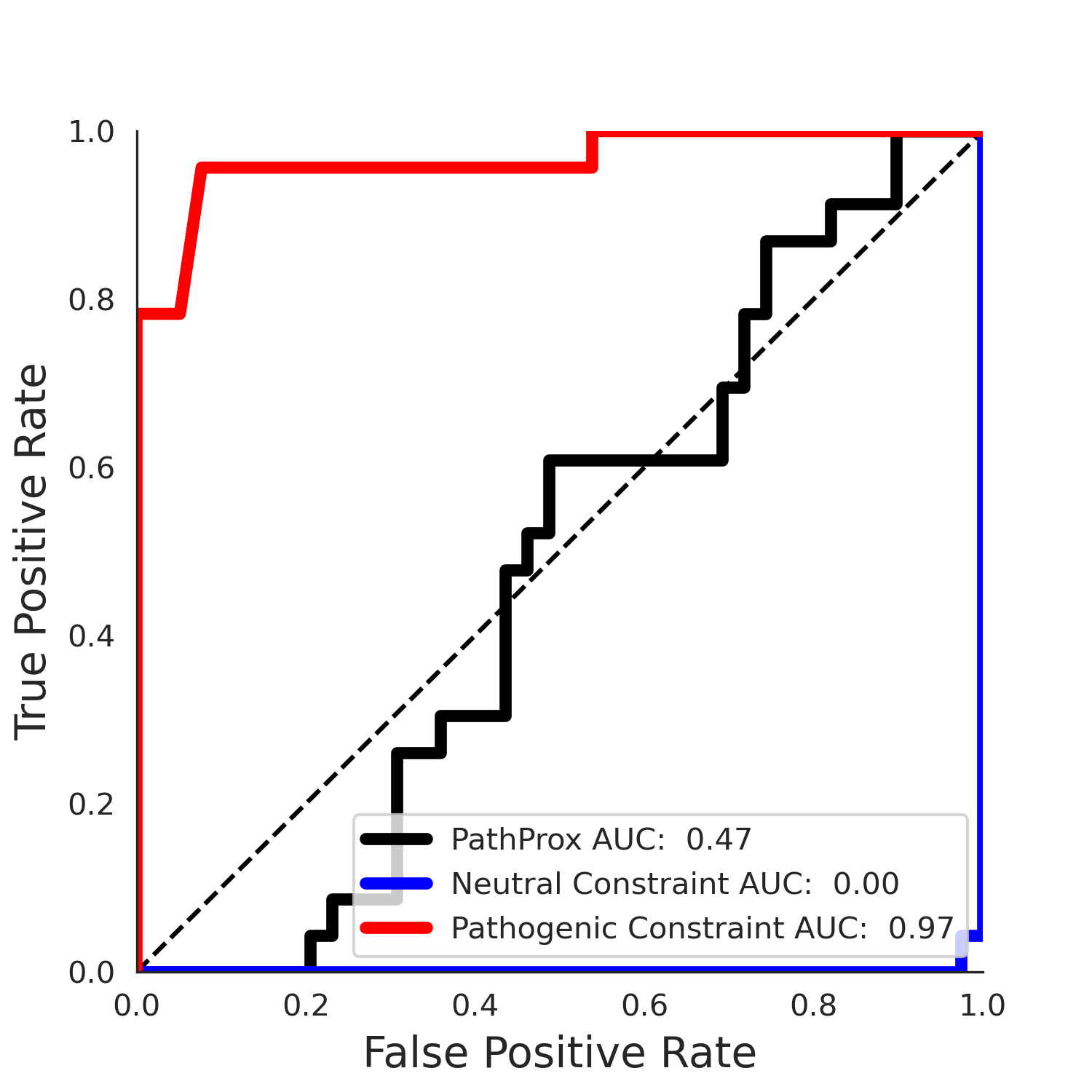
PathProx clinvar PR
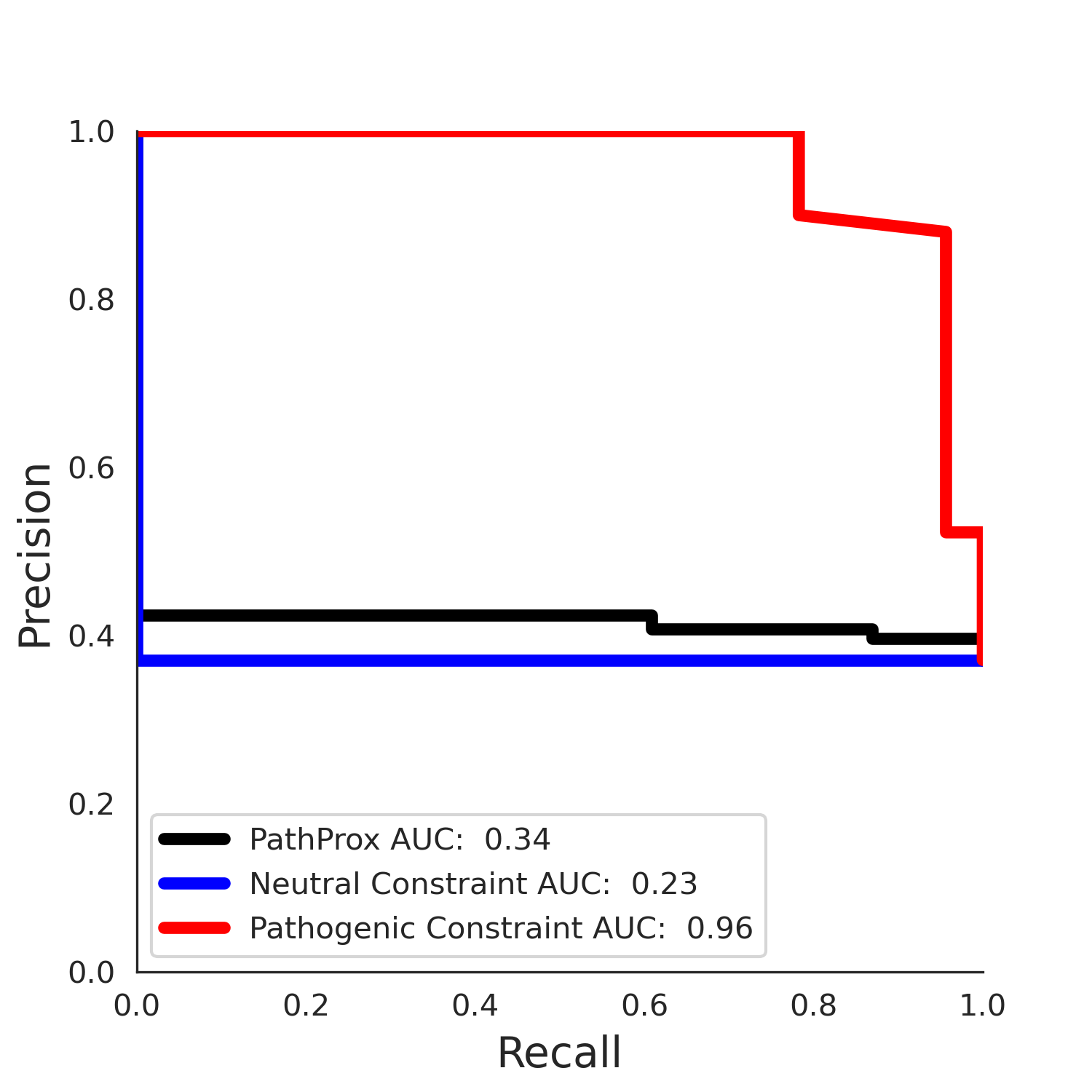
PathProx (COSMIC - gnomAD38) Results
PathProx

PathProx COSMIC ROC
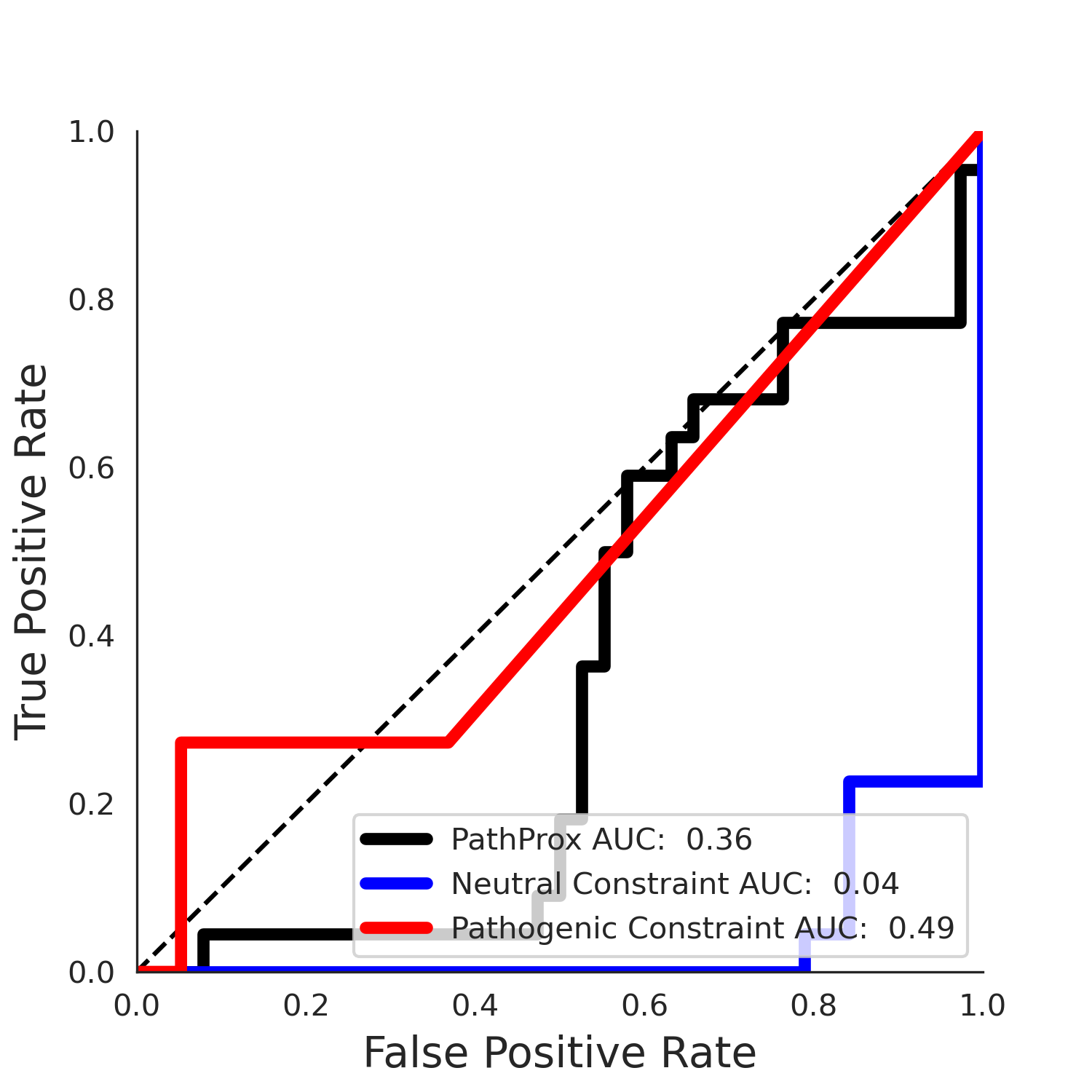
PathProx COSMIC PR
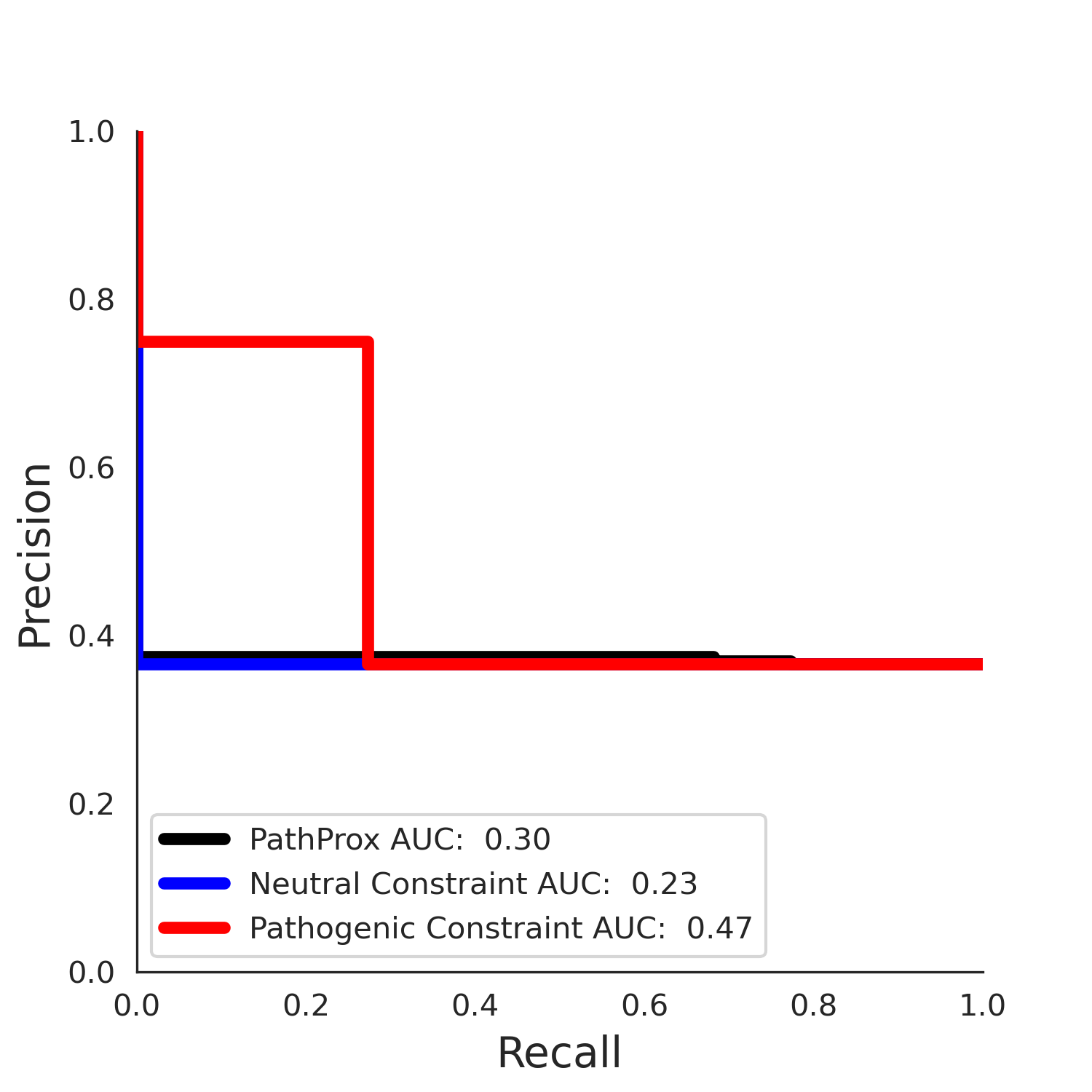
ENSP00000337131.4_2.A monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| modbase | ENSP00000337131.4_2 | A | 5txt | 37.0 | 1.0 | 106 | 510 | 0.37 | 0.908 | 0.03 | 0.498 |
clinvar Results - ENSP00000337131.4_2.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
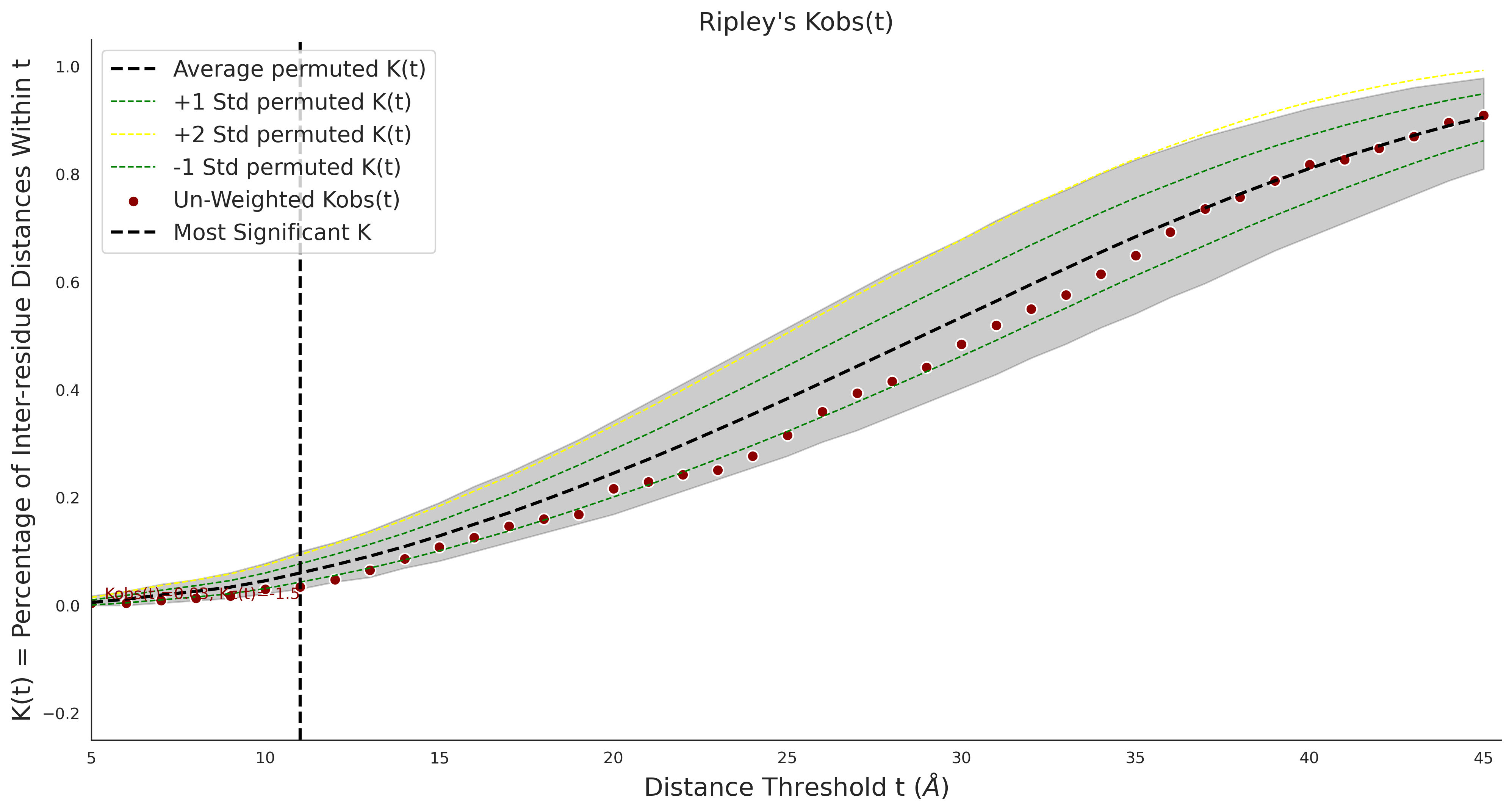
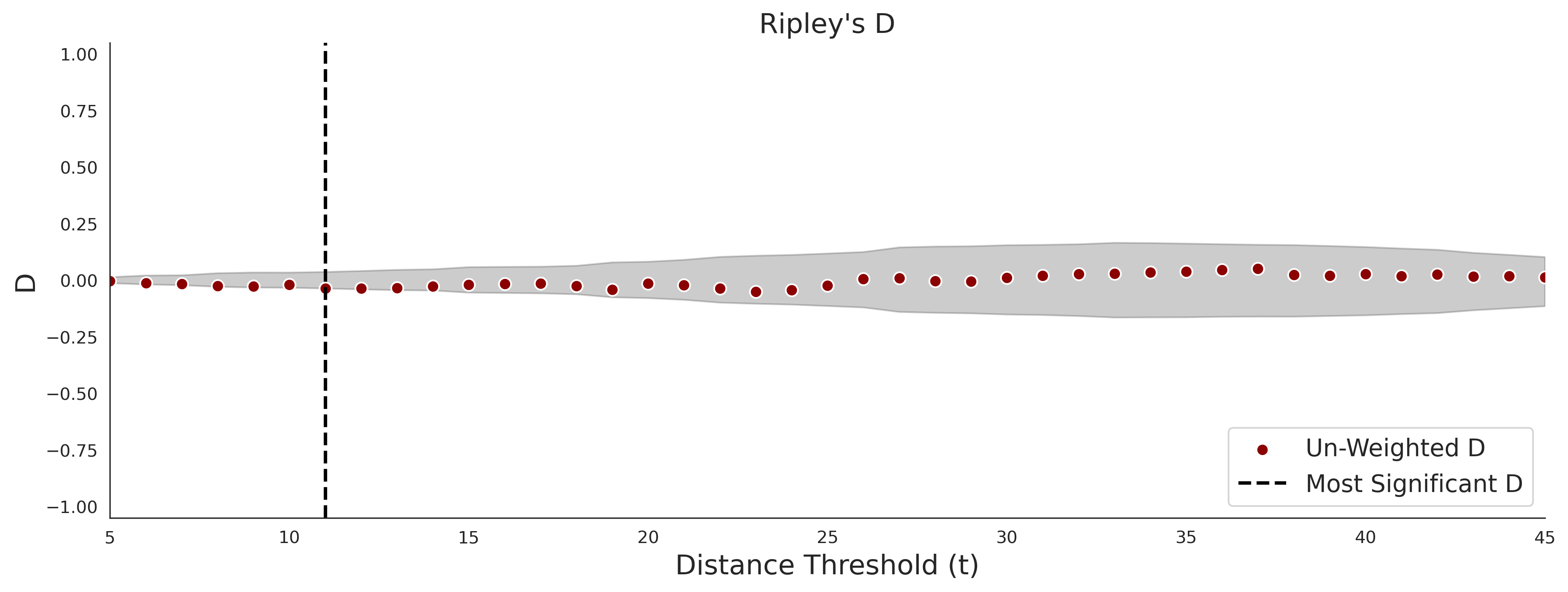
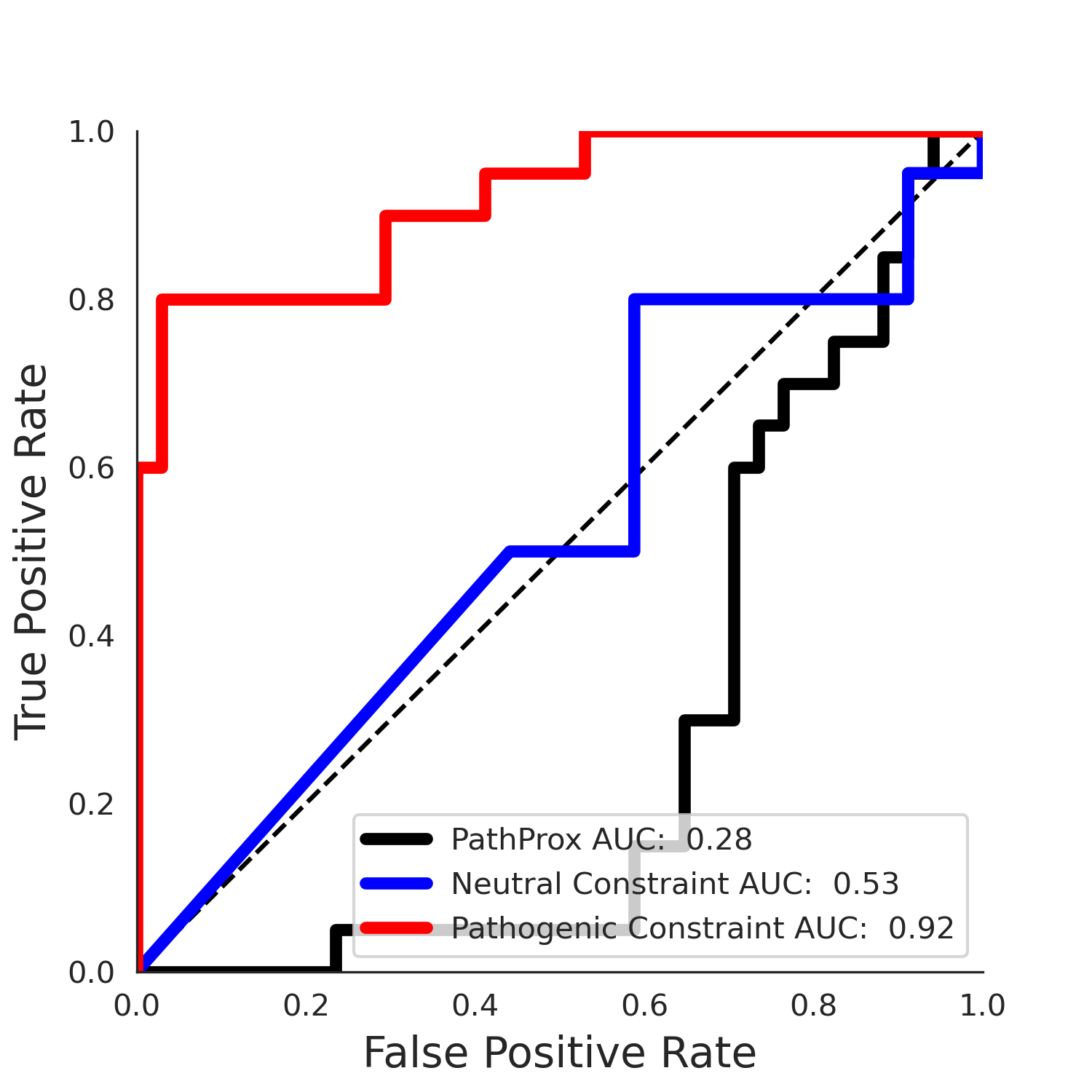
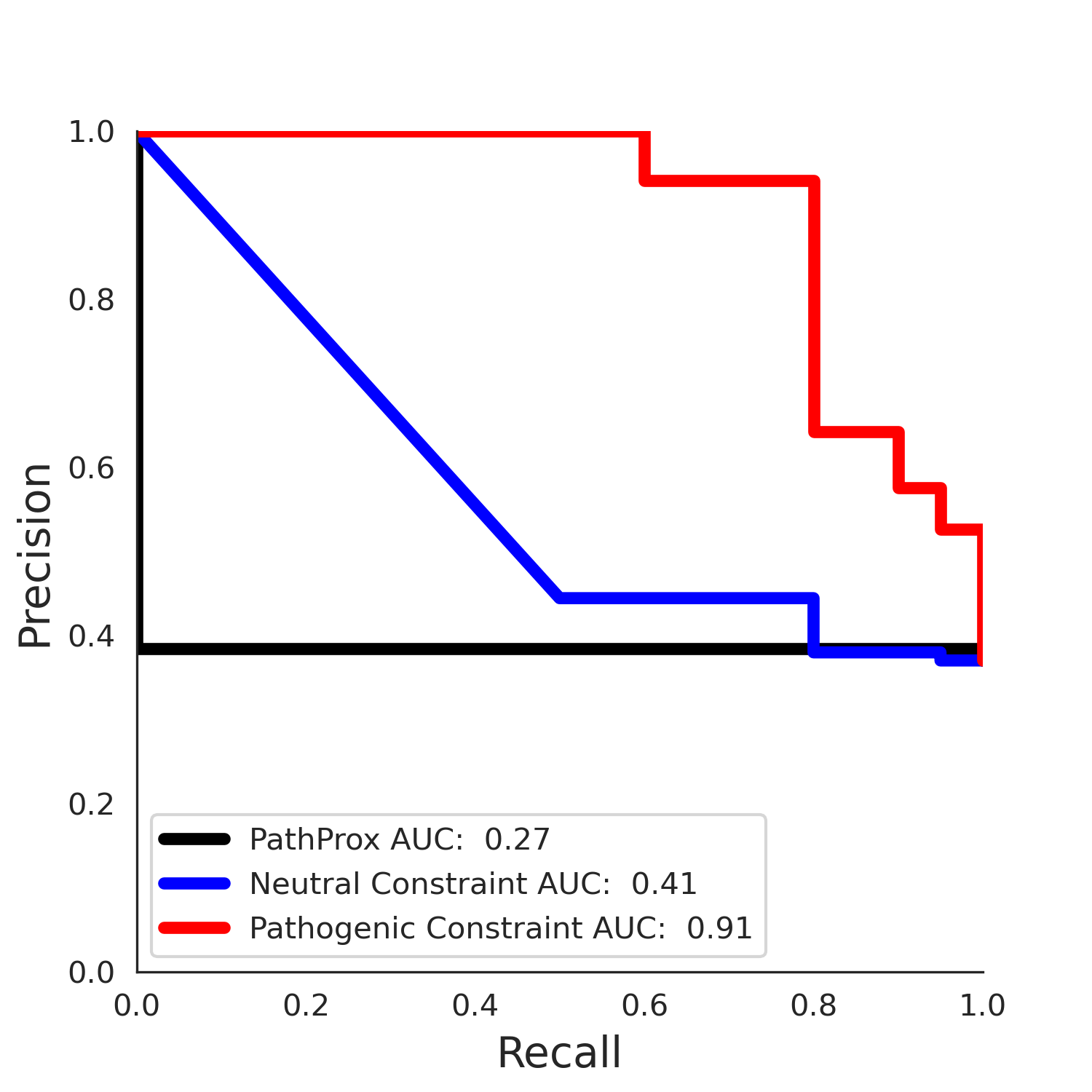
| 1.71 | 8.00 | 1.14 | 37.00 | 0.92 | 0.91 | 0.37 | 0.81 | 0.32 |
COSMIC Results - ENSP00000337131.4_2.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| 1.71 | 8.00 | -1.49 | 11.00 | 0.45 | 0.50 | 0.03 | 1.00 | 0.02 |
Location of A245T in ENSP00000337131.4_2.A (NGL Viewer)
Variant Distributions
33 gnomAD38
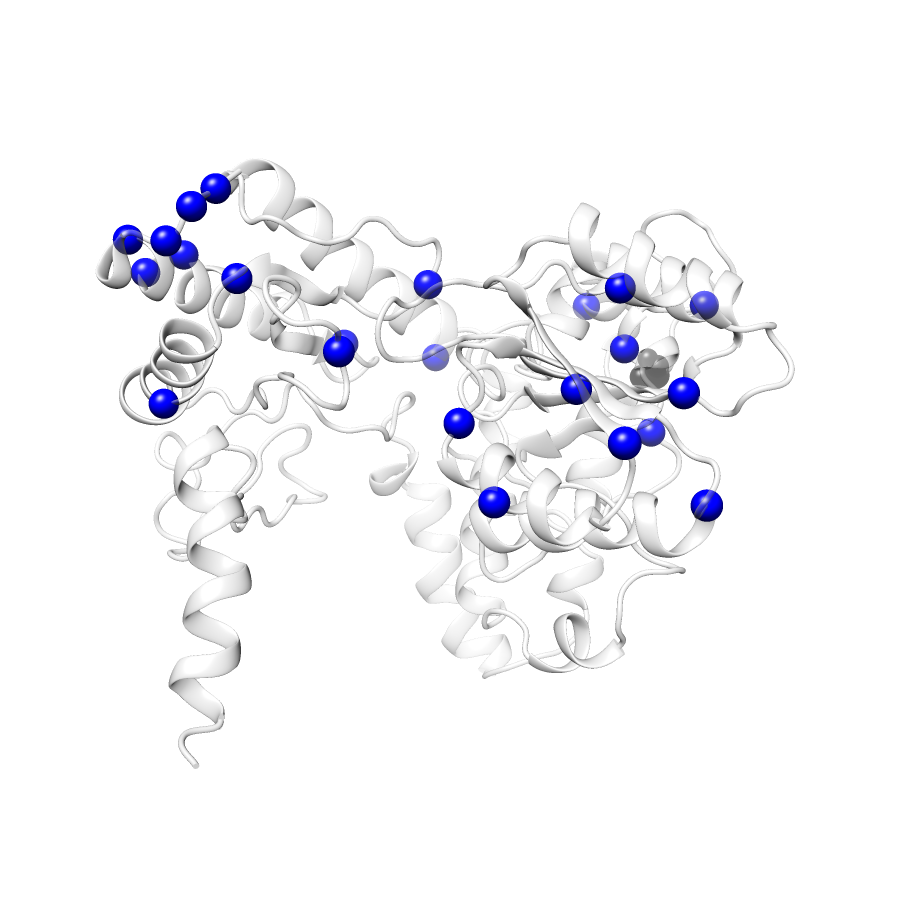
20 clinvar
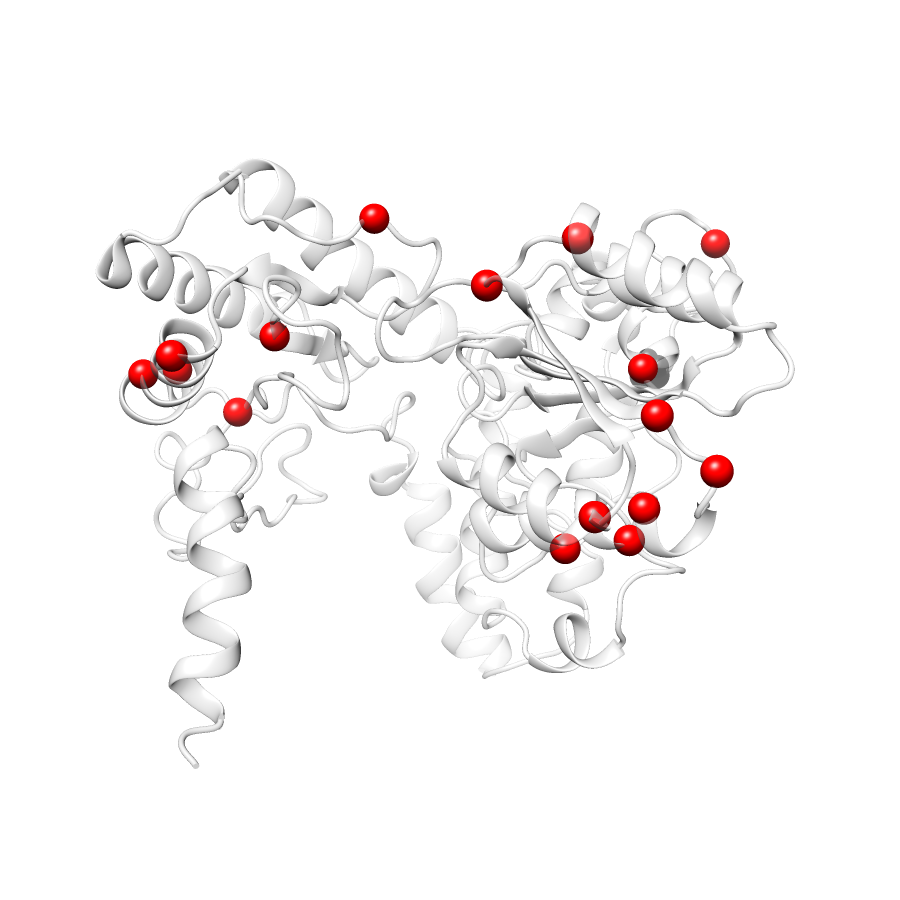
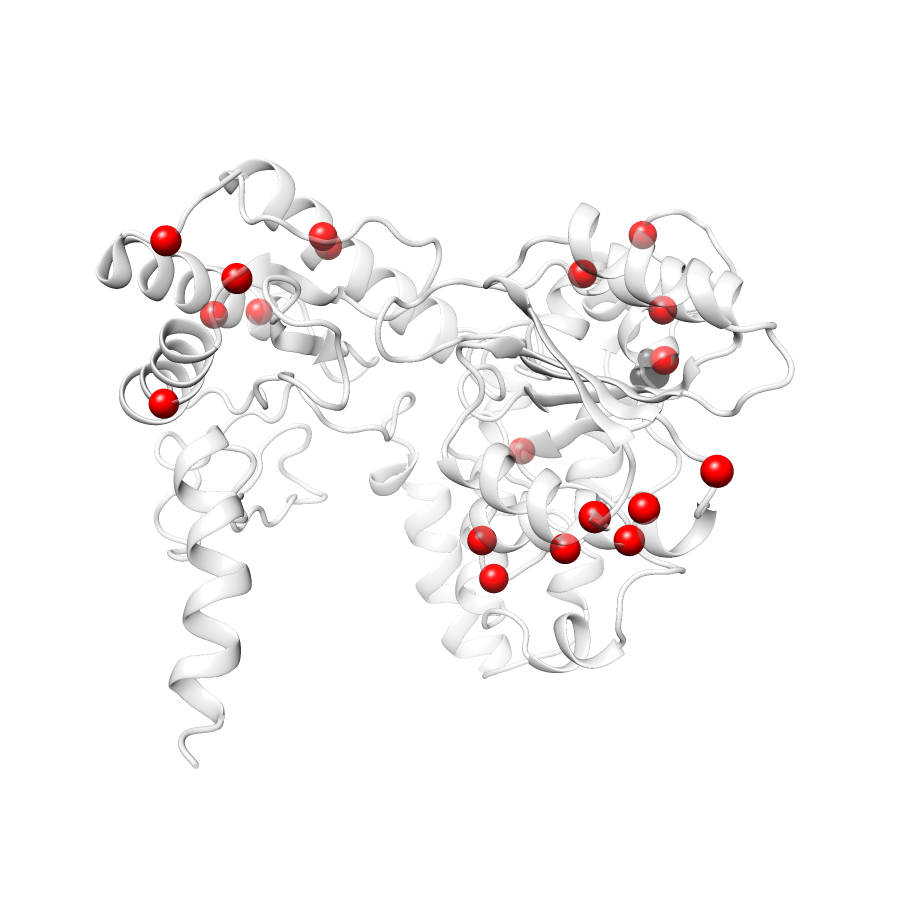
22 COSMIC
Ripley's Univariate K
gnomAD38
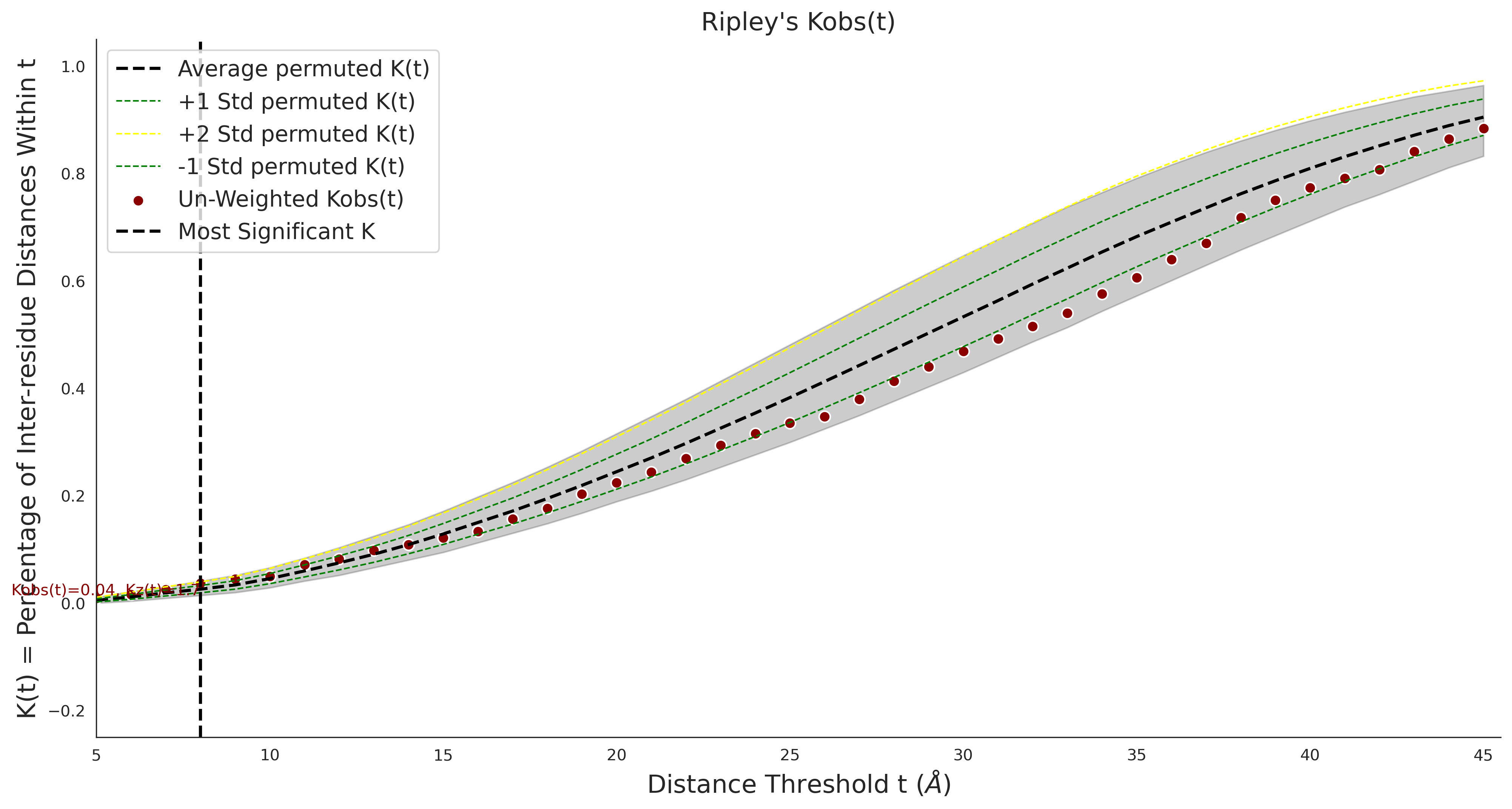
clinvar
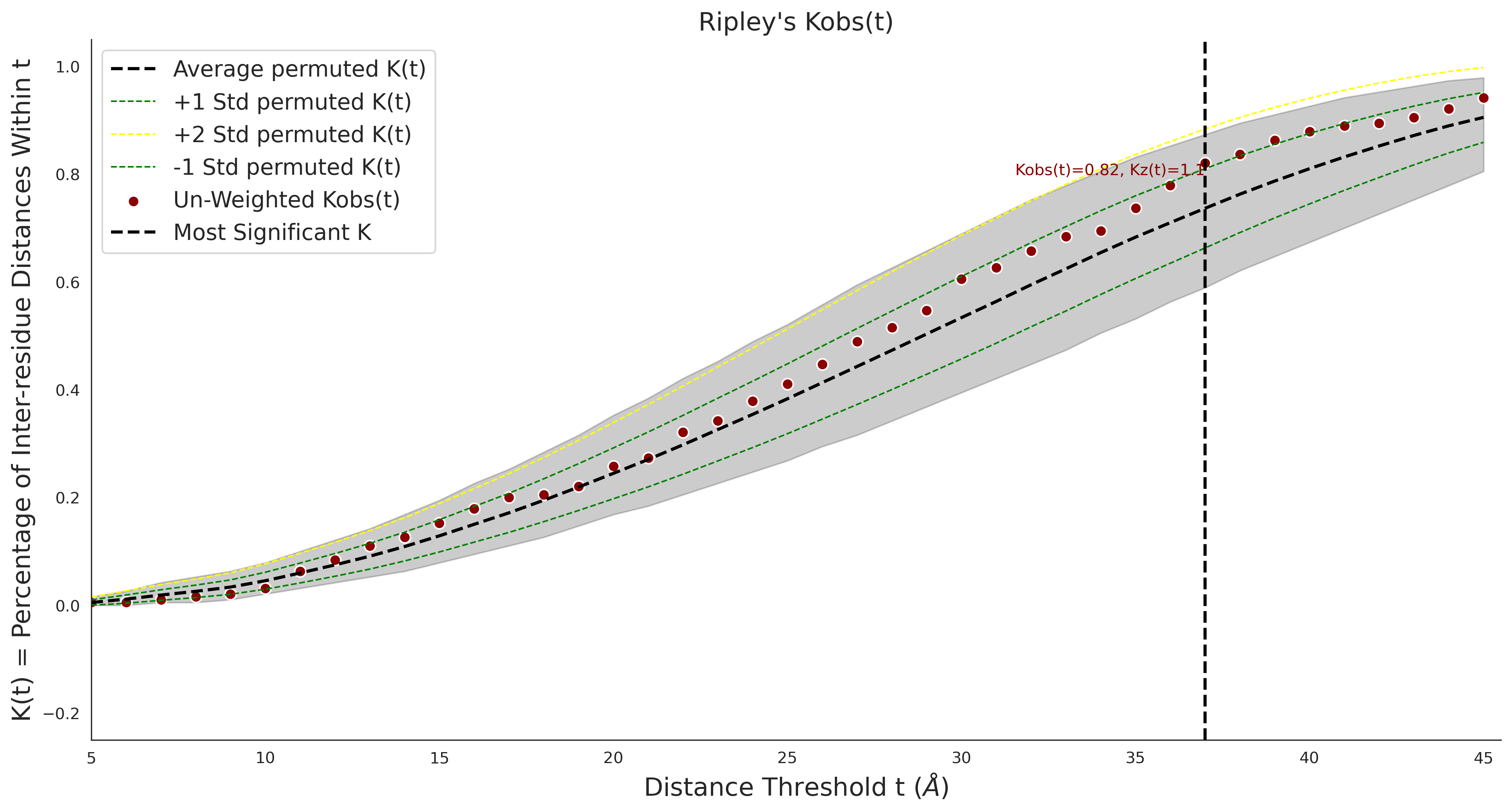
COSMIC
Ripley's Bivariate D
clinvar - gnomAD38
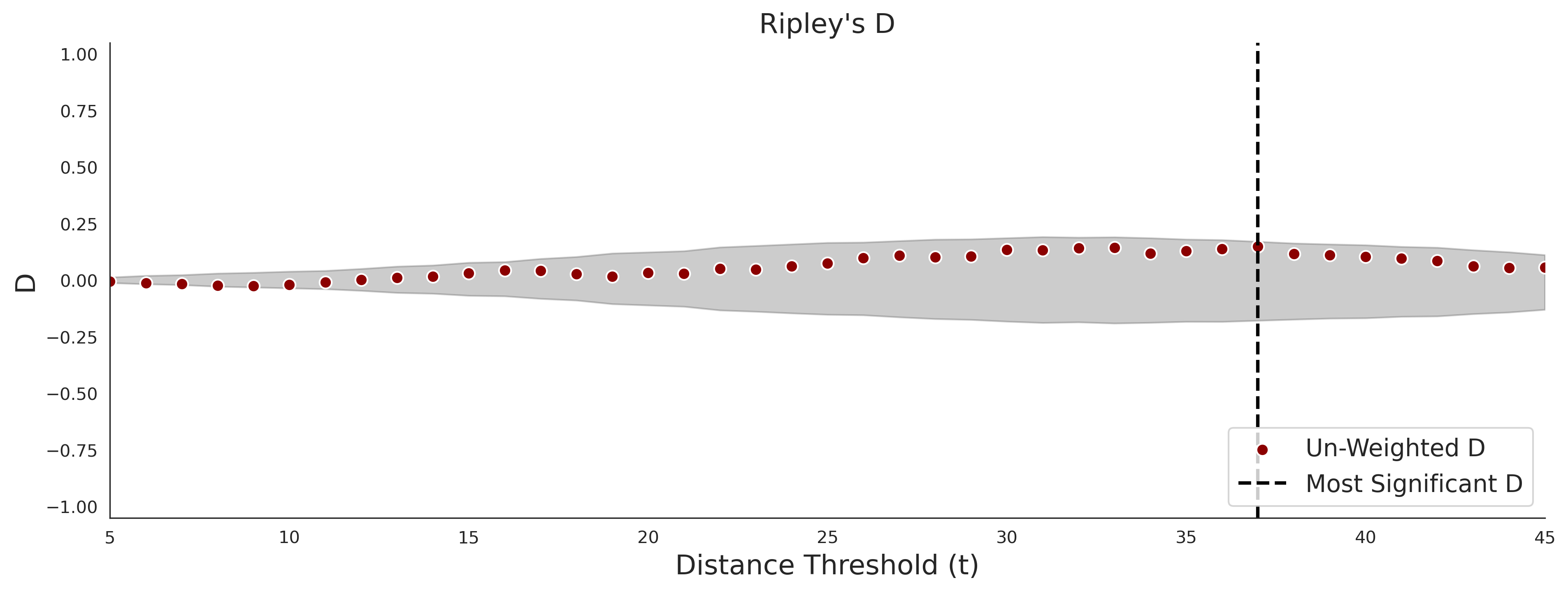
COSMIC - gnomAD38
PathProx (clinvar - gnomAD38) Results
PathProx
PathProx clinvar ROC
PathProx clinvar PR
PathProx (COSMIC - gnomAD38) Results
PathProx
PathProx COSMIC ROC
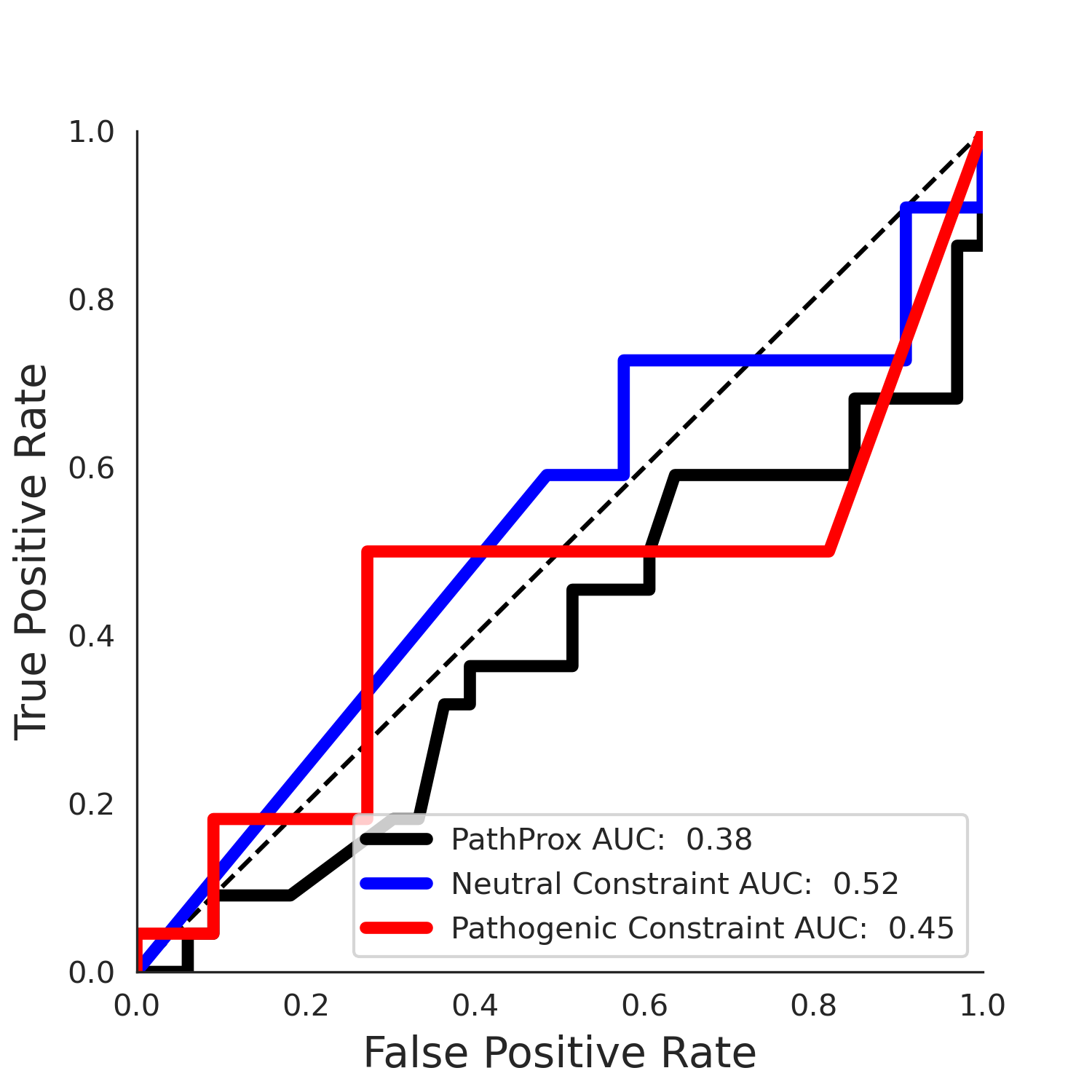
PathProx COSMIC PR
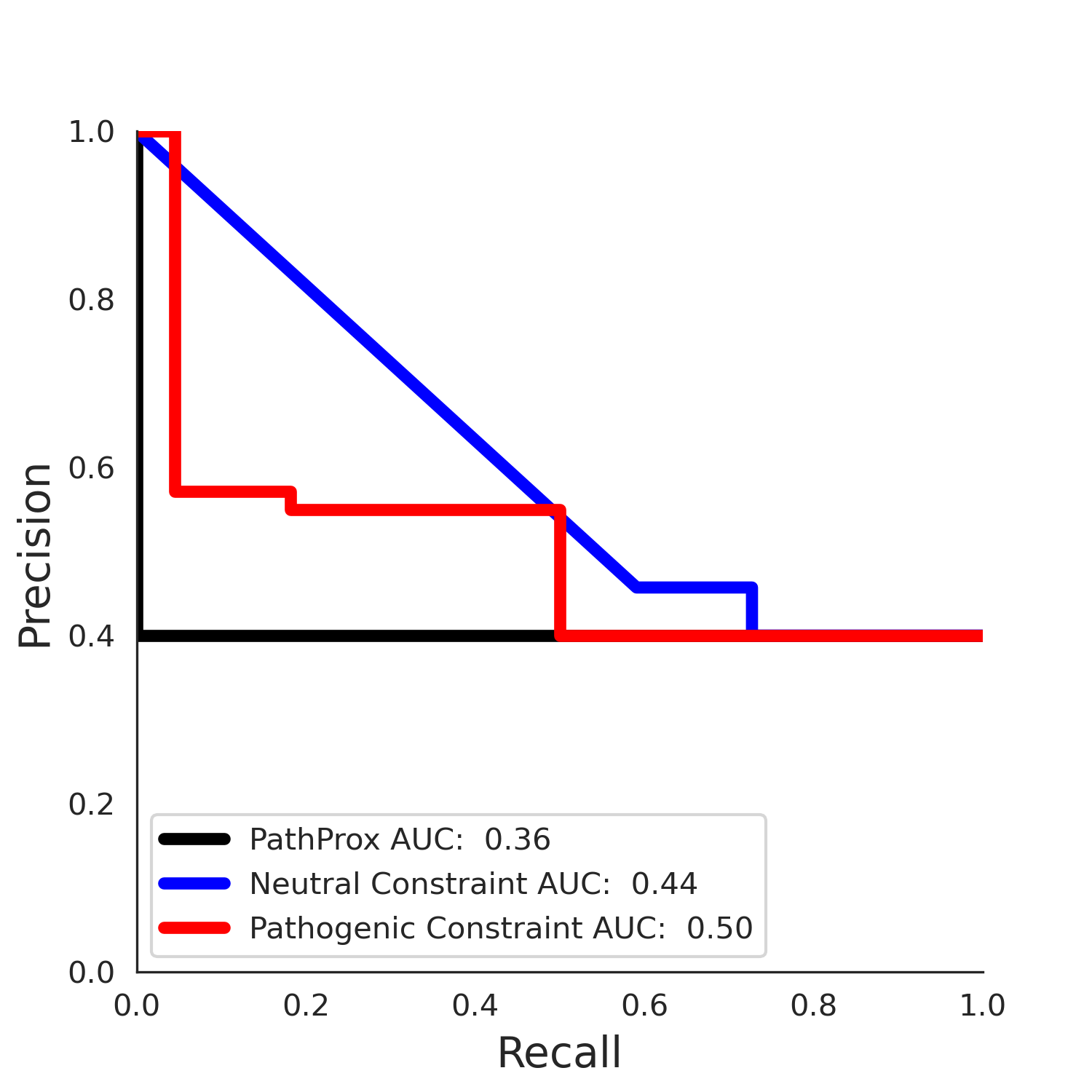
ENSP00000337131.4_3.A monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| modbase | ENSP00000337131.4_3 | A | 2bwn | 50.0 | 1.0 | 107 | 503 | 2.3 | 2.97 | 0.29 | 0.874 | -0.02 | 0.479 |
clinvar Results - ENSP00000337131.4_3.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -1.98 | 25.00 | 2.51 | 26.00 | 0.86 | 0.87 | 0.29 | 0.92 | 0.18 |
COSMIC Results - ENSP00000337131.4_3.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -1.94 | 27.00 | -1.61 | 9.00 | 0.44 | 0.48 | -0.02 | 1.00 | 0.00 |
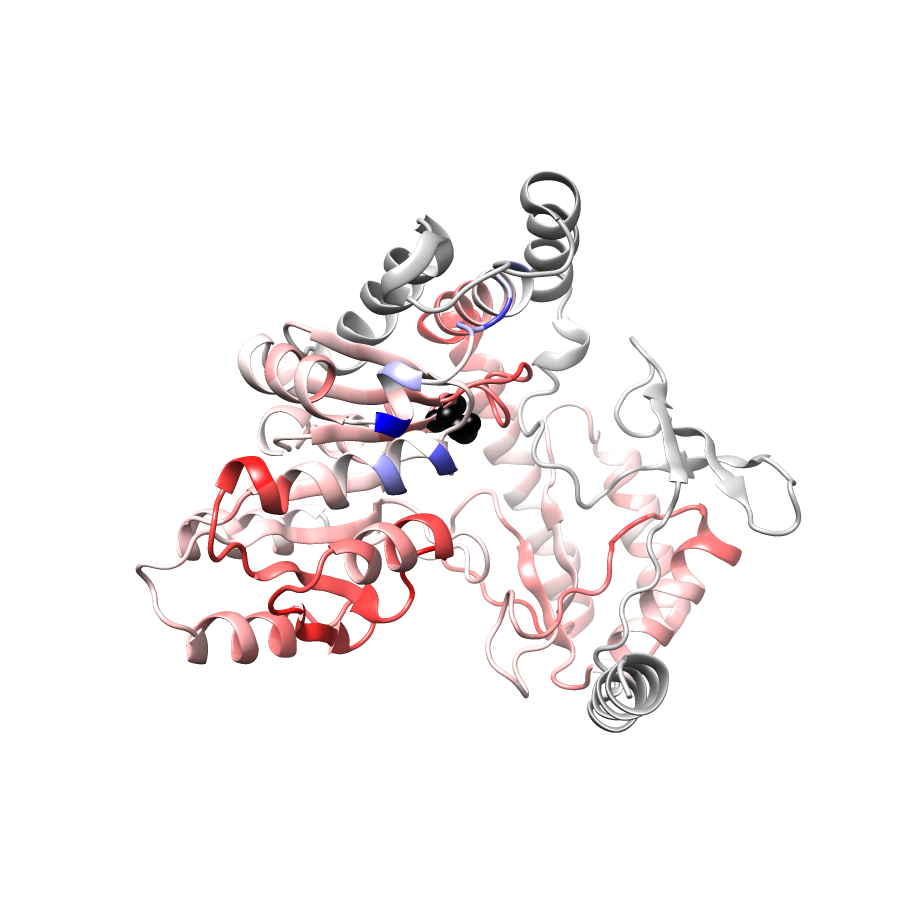
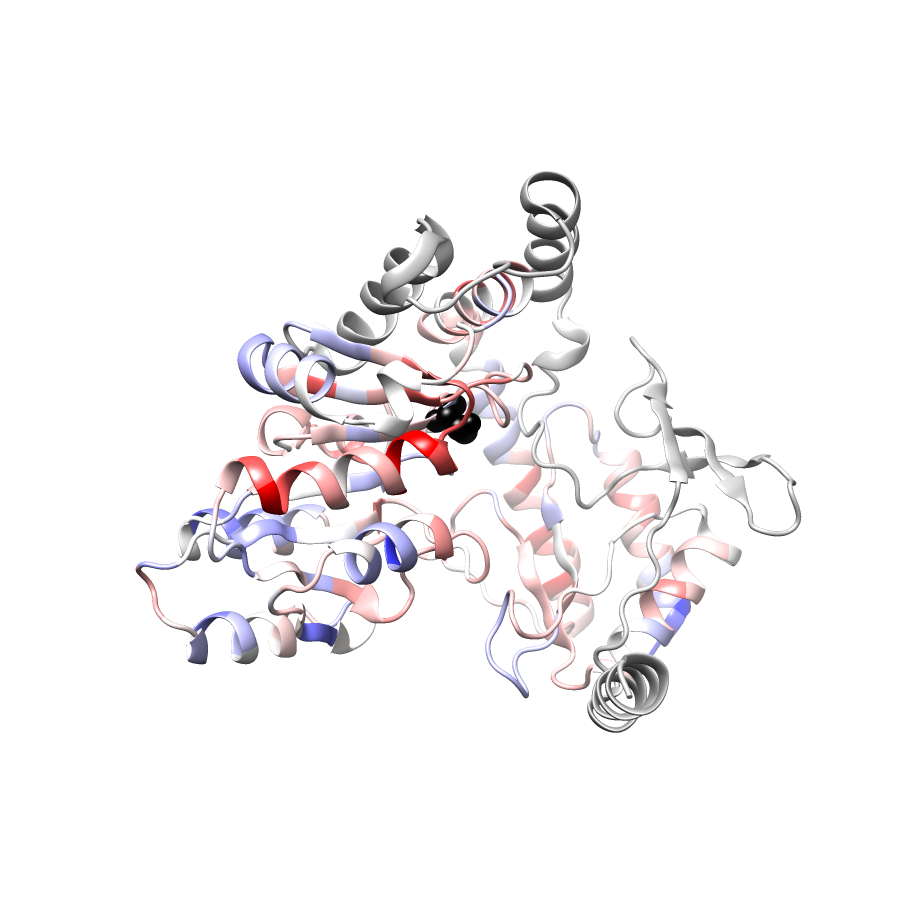
Location of A245T in ENSP00000337131.4_3.A (NGL Viewer)
Variant Distributions
31 gnomAD38
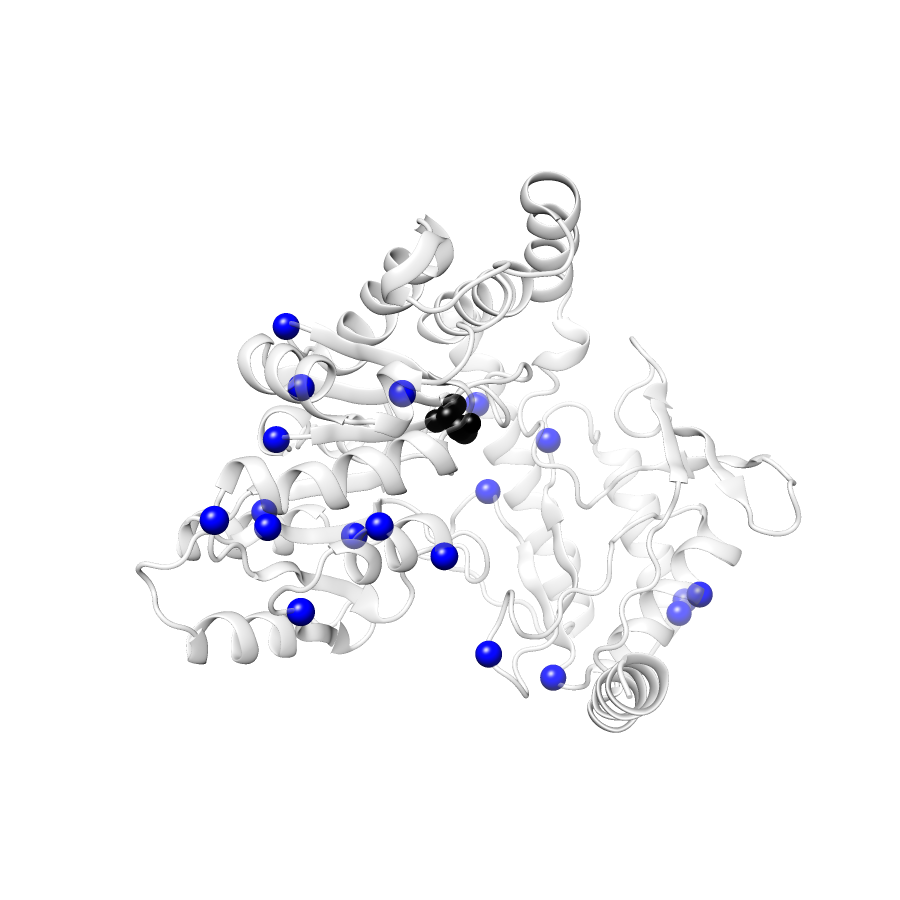
20 clinvar
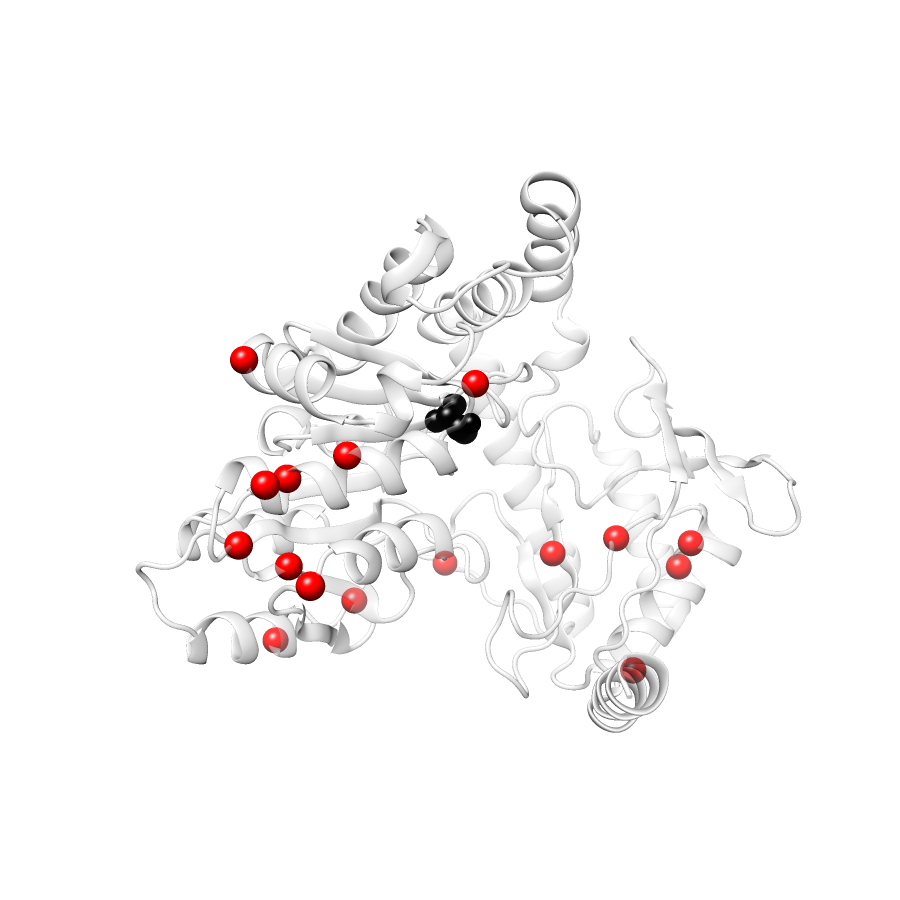
21 COSMIC
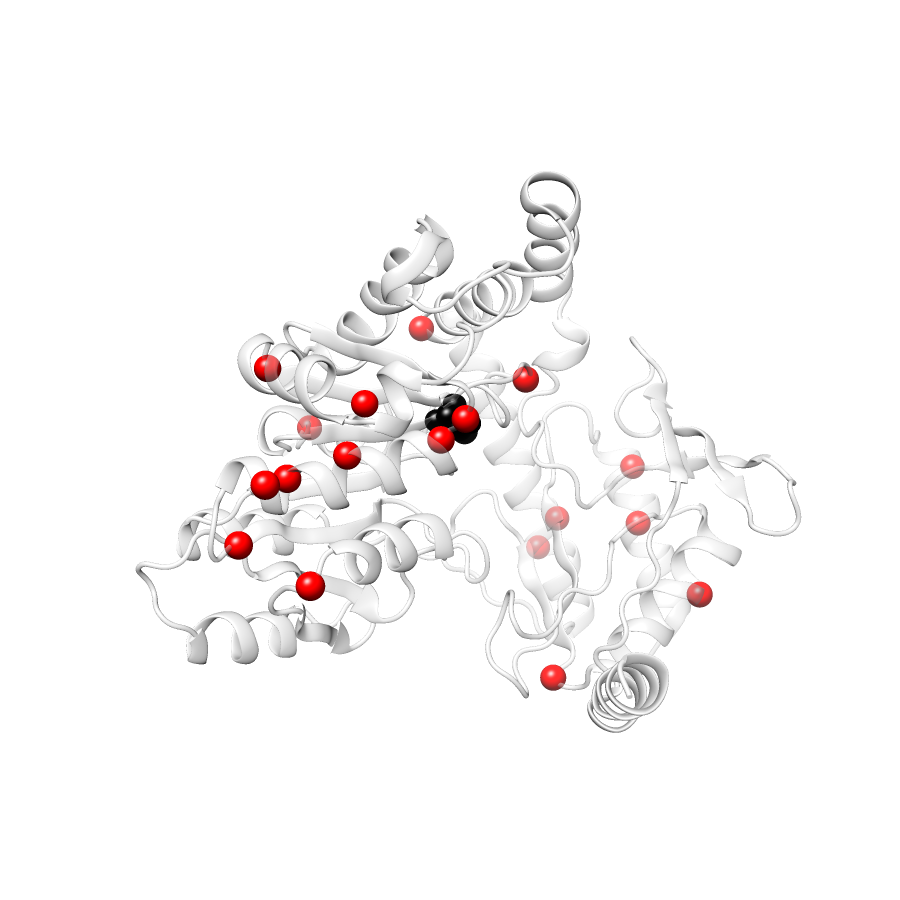
Ripley's Univariate K
gnomAD38
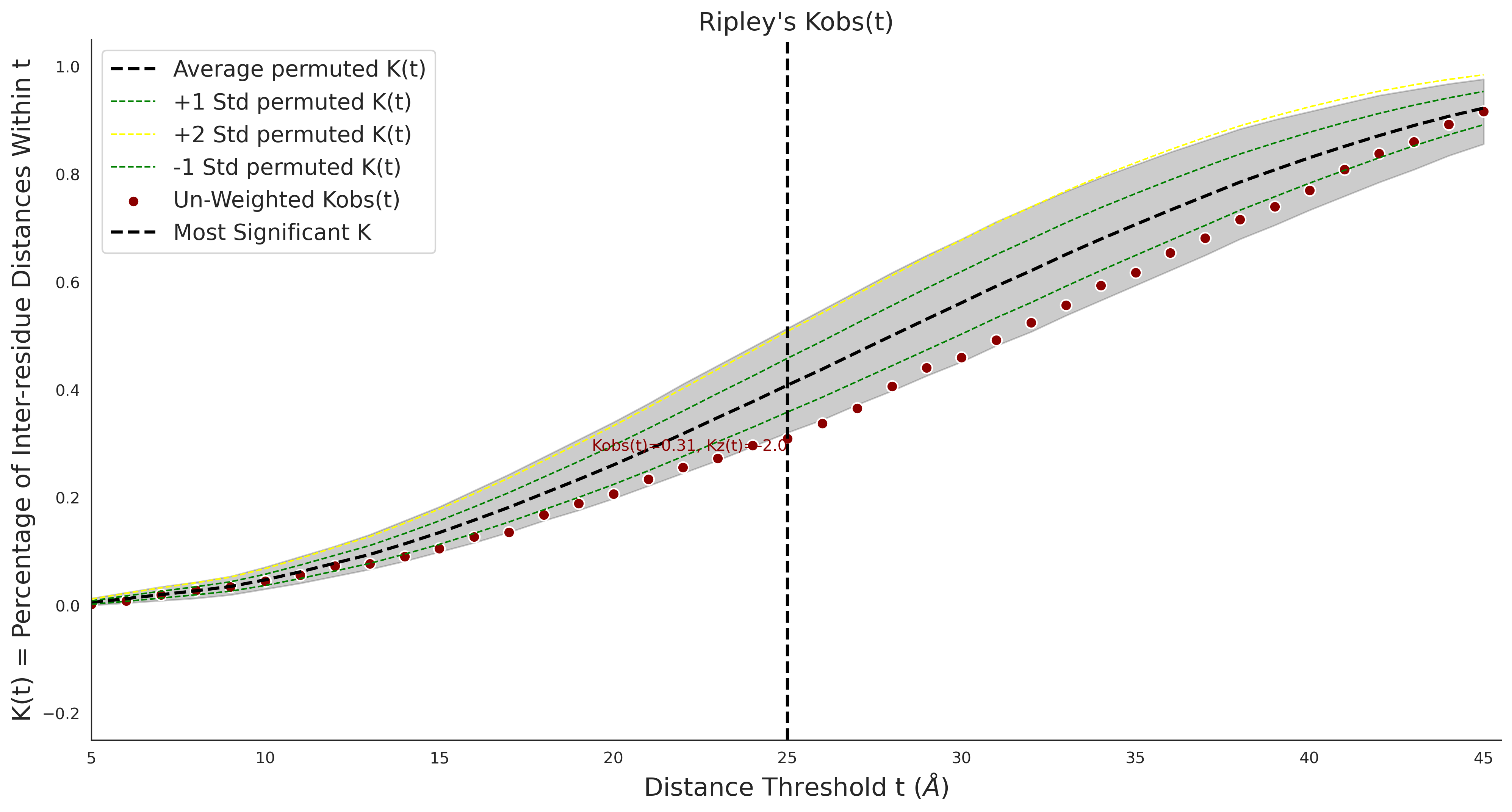
clinvar
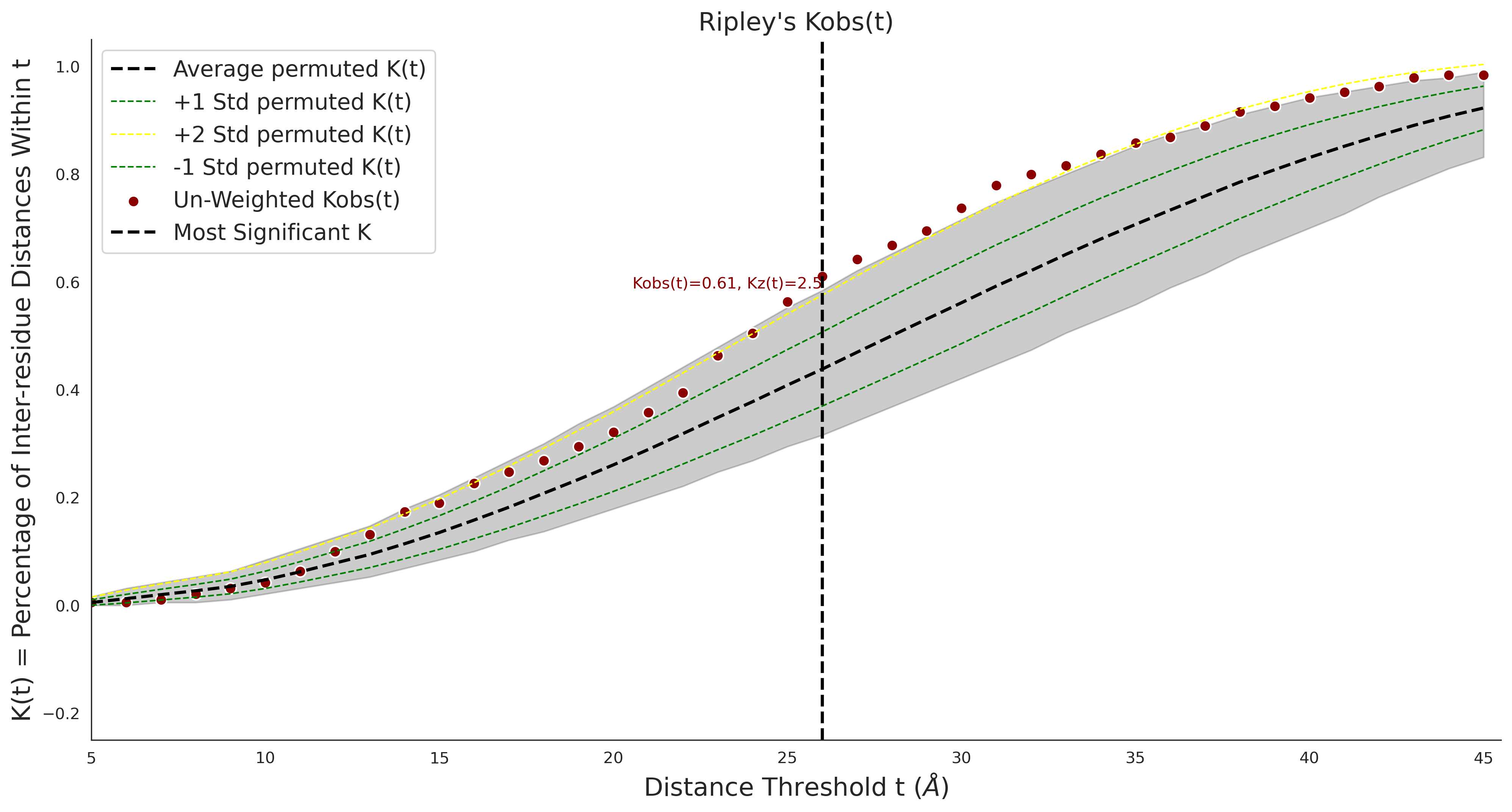
COSMIC
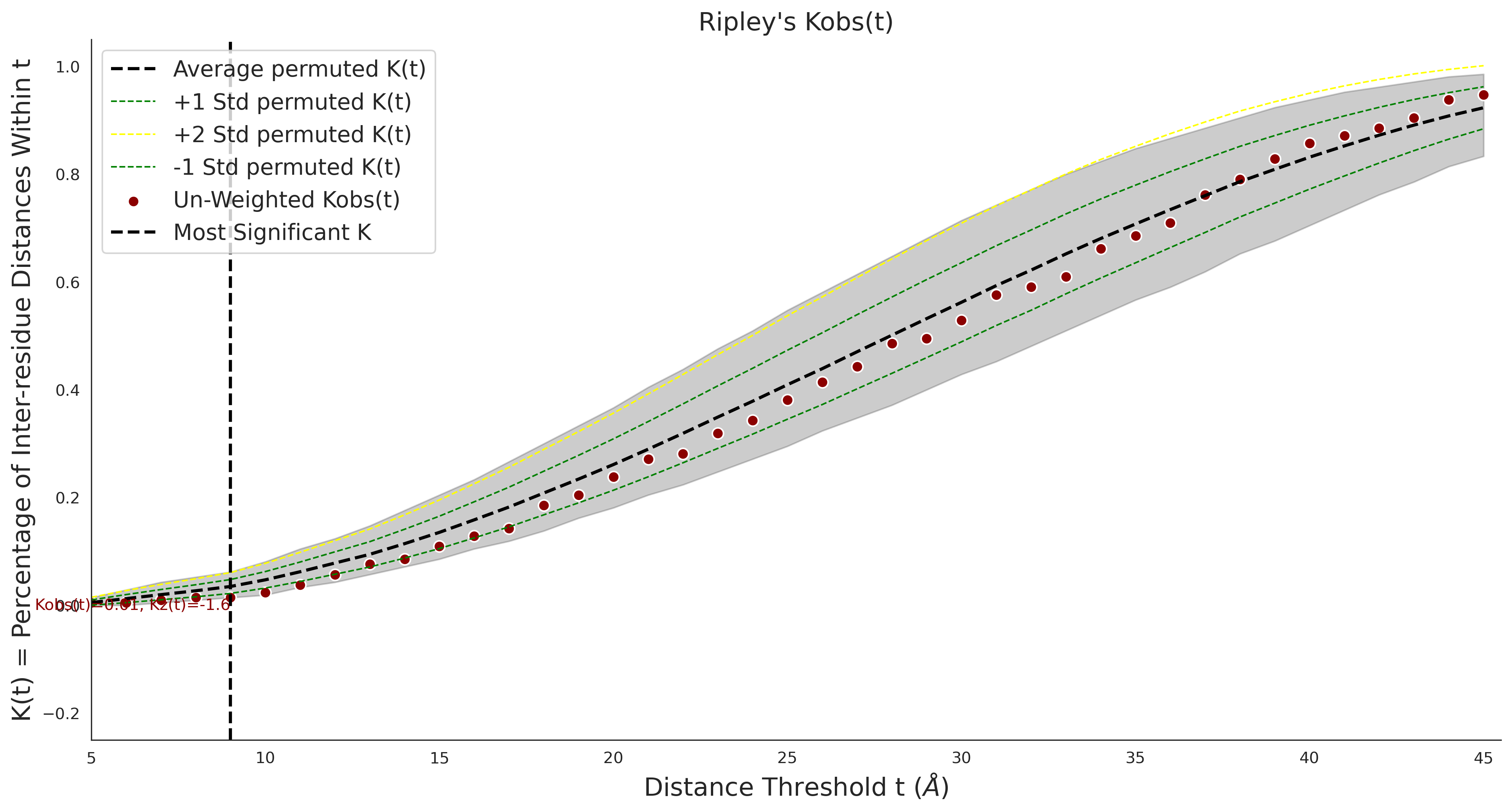
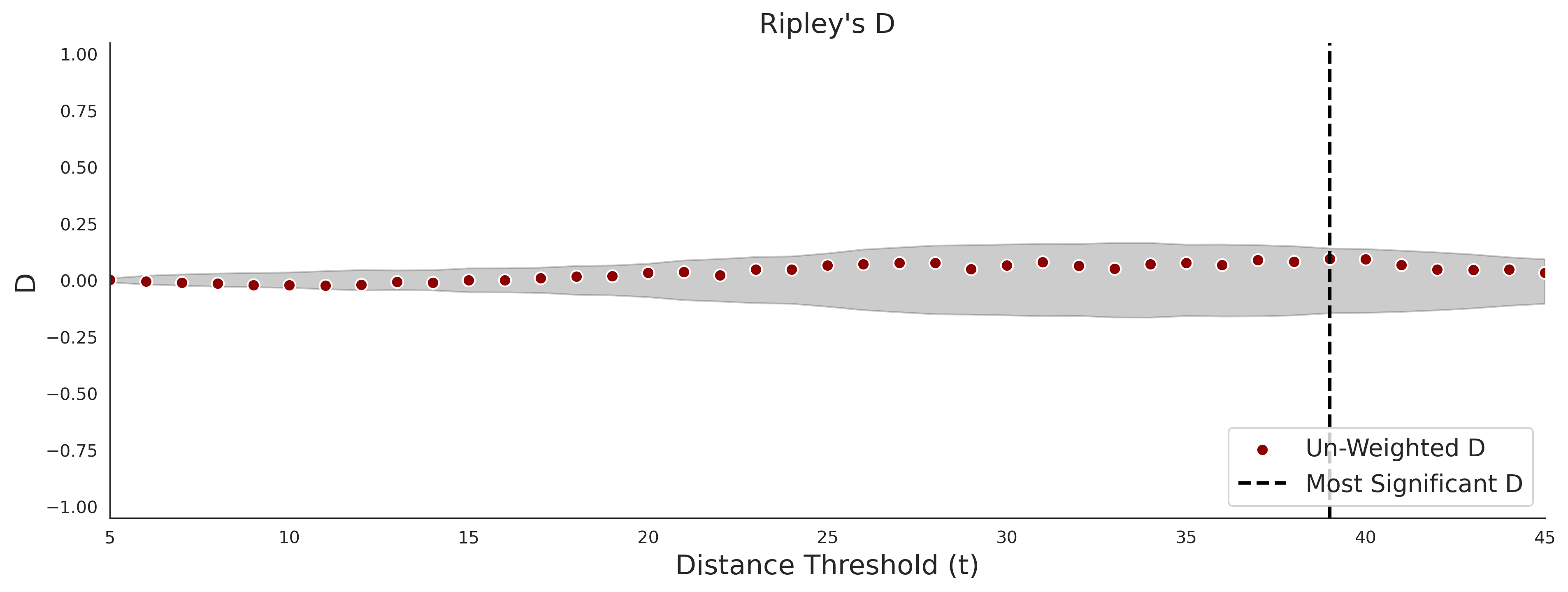
Ripley's Bivariate D
clinvar - gnomAD38
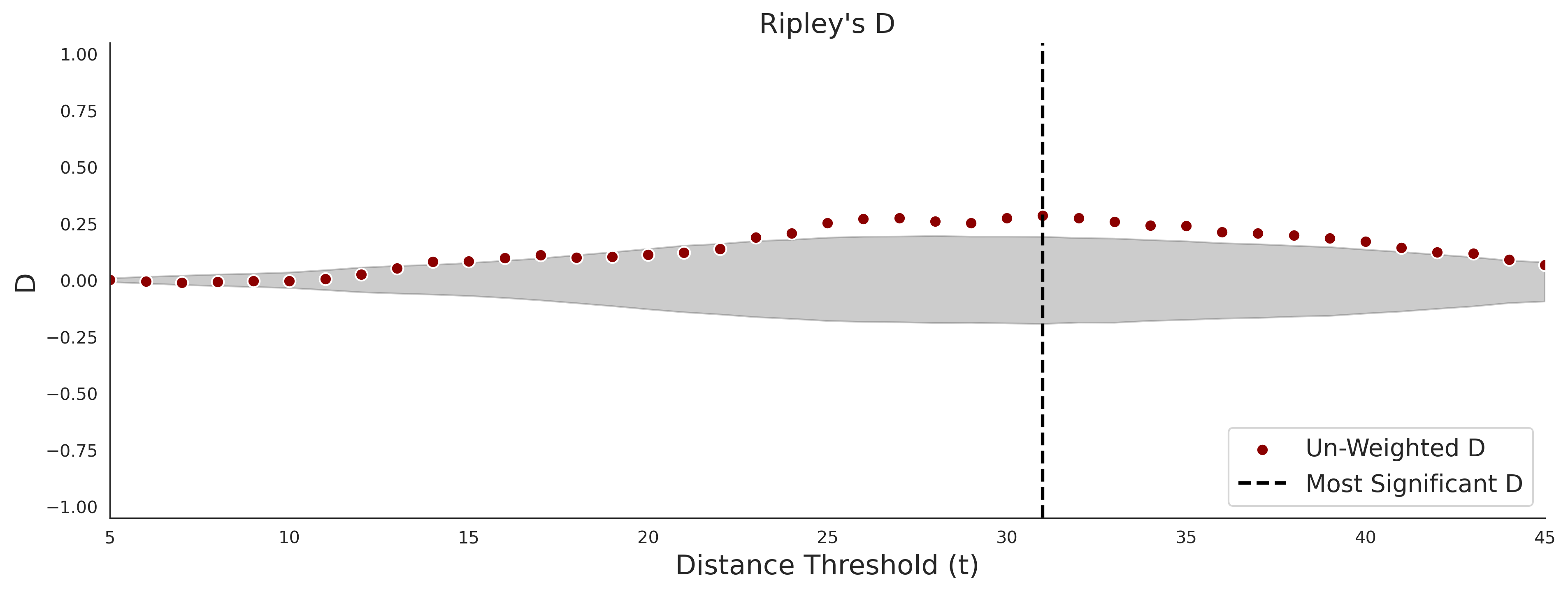
COSMIC - gnomAD38
PathProx (clinvar - gnomAD38) Results
PathProx
PathProx clinvar ROC
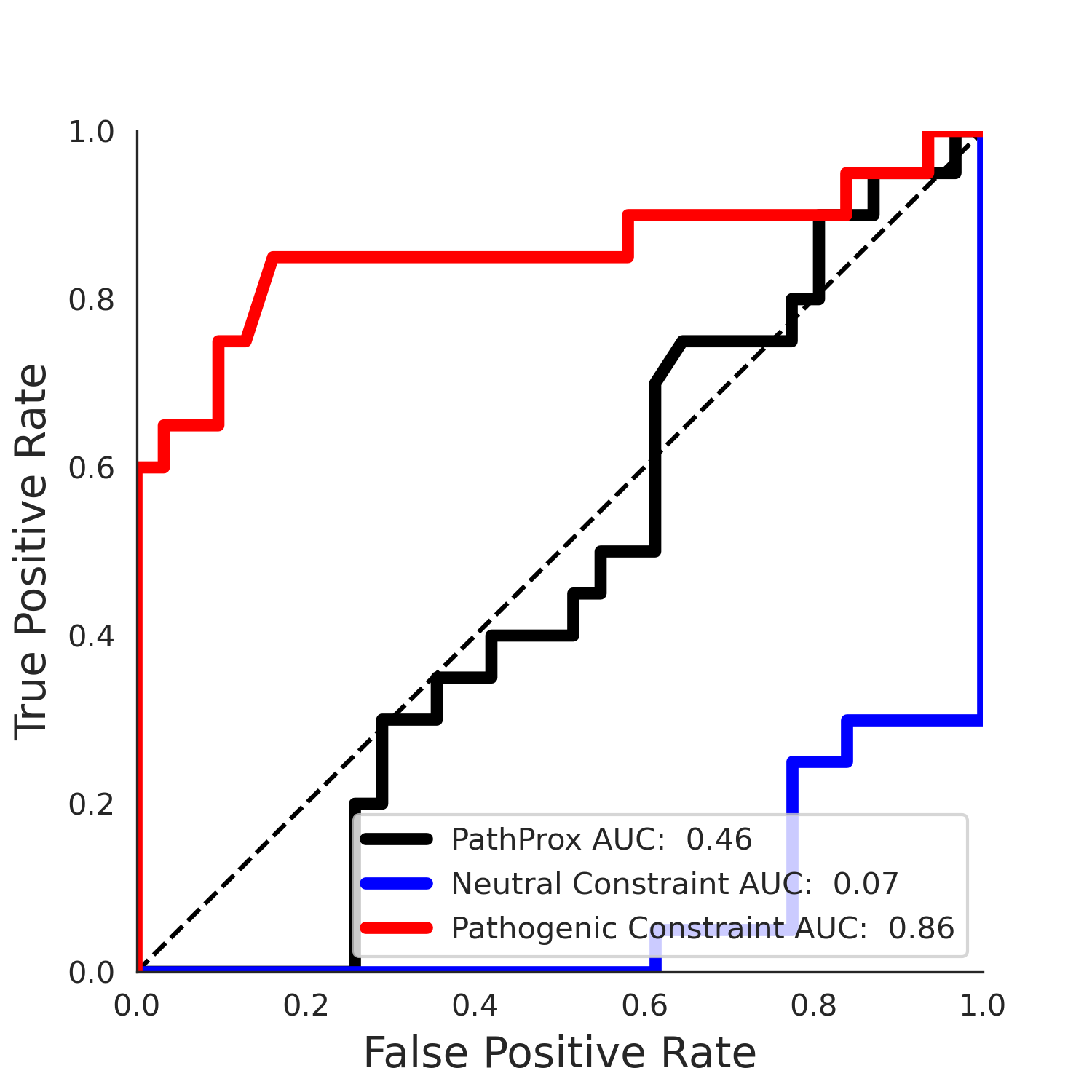
PathProx clinvar PR
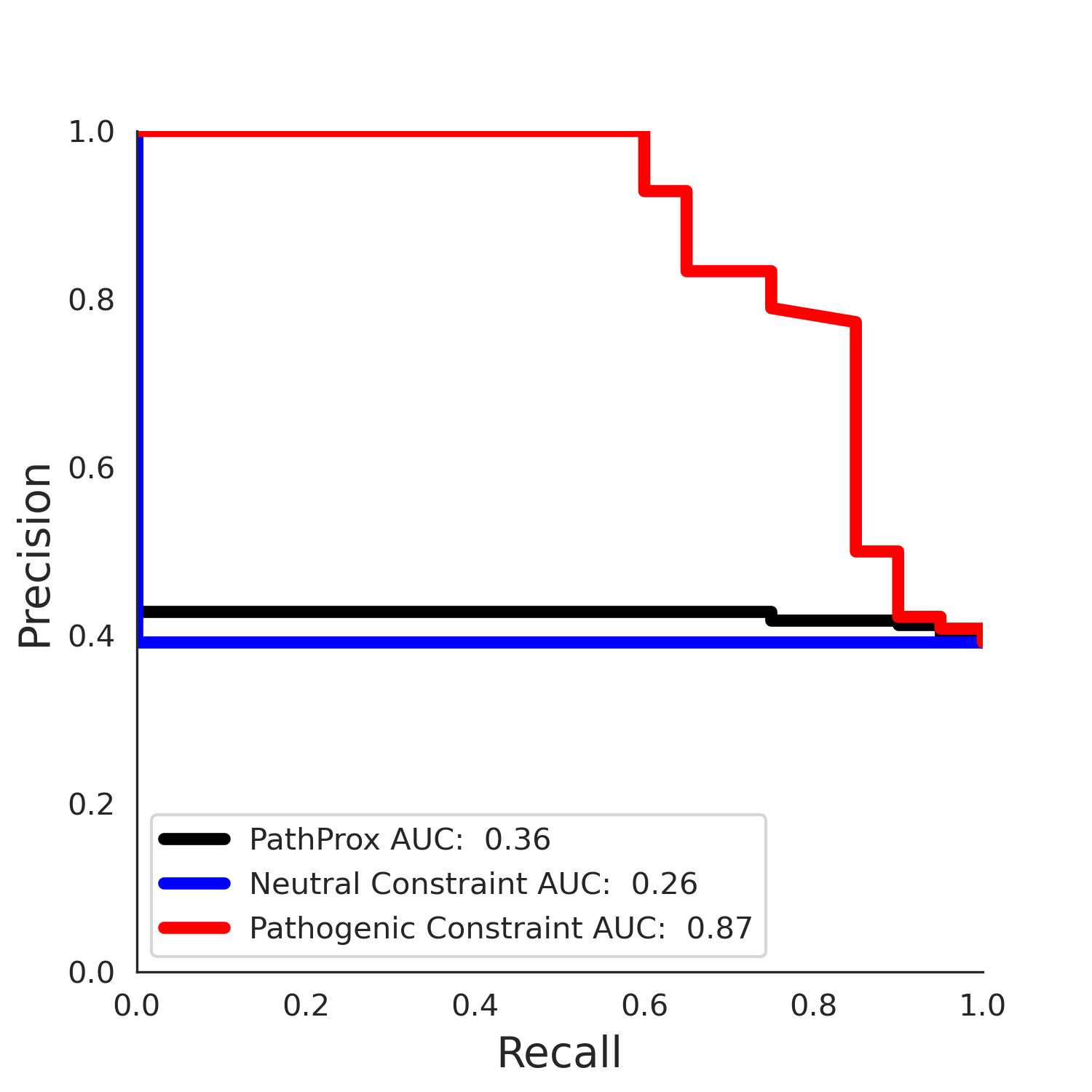
PathProx (COSMIC - gnomAD38) Results
PathProx
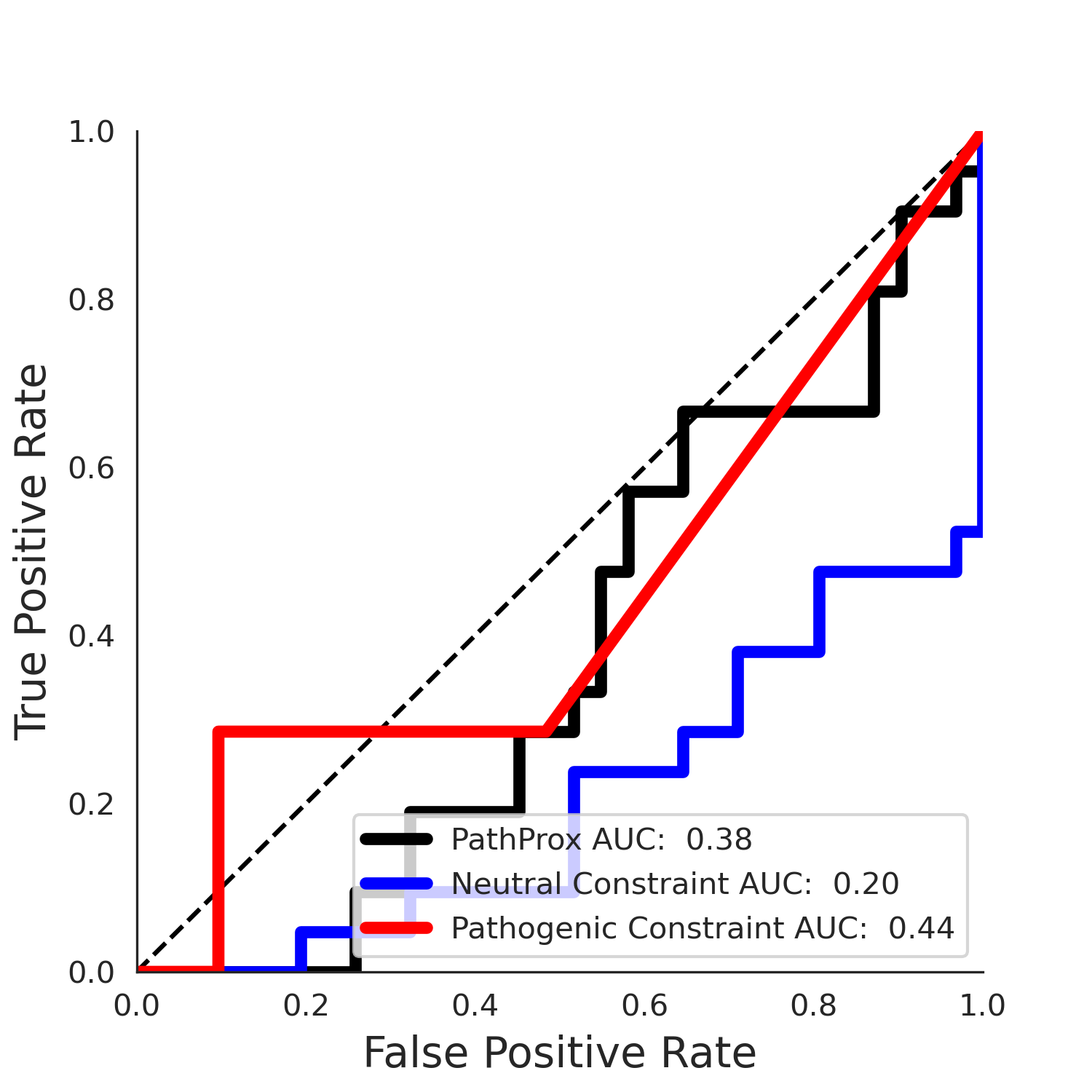
PathProx COSMIC ROC
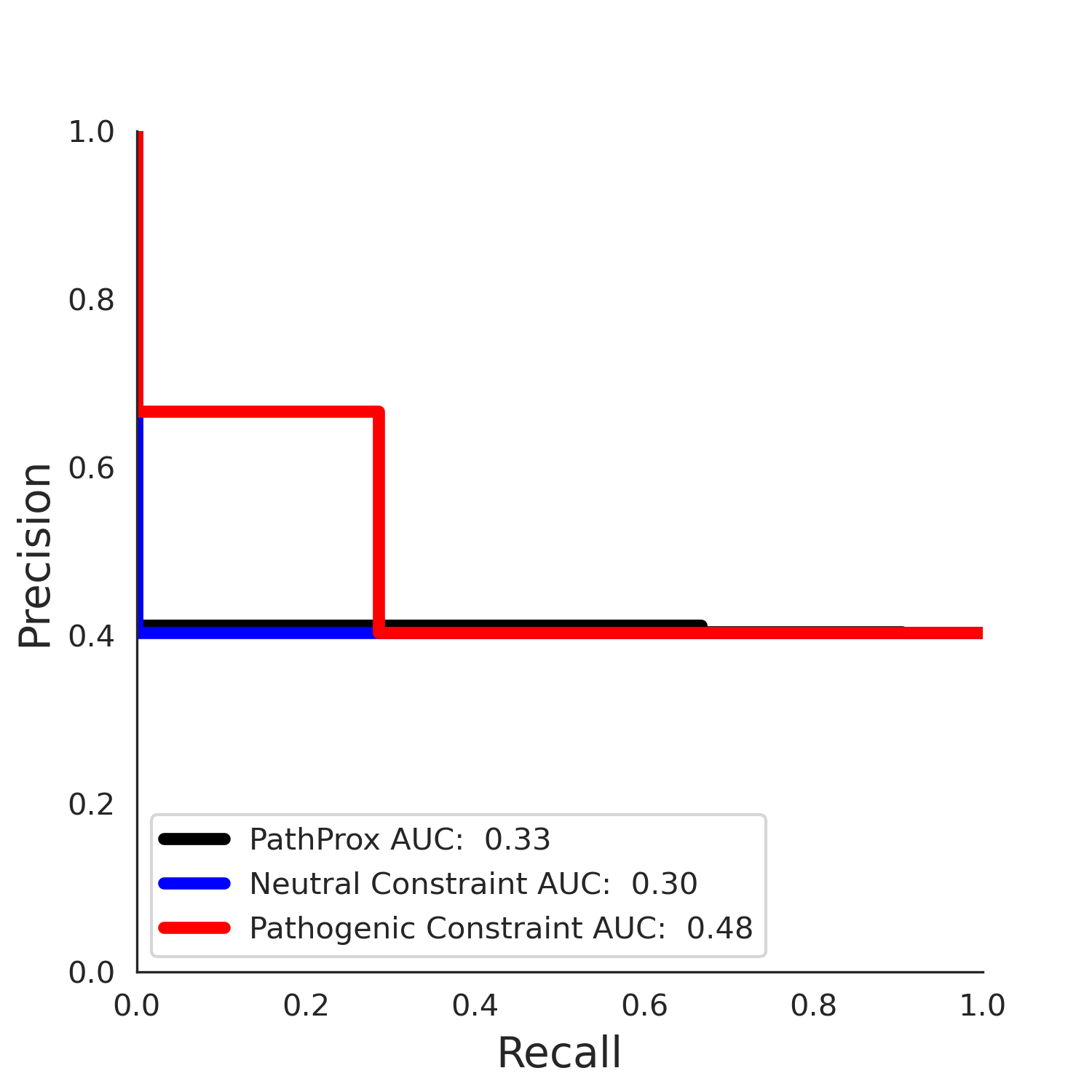
PathProx COSMIC PR
5qqr.A dimer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| X-RAY | 5qqr | A | 1.46 | 1.0 | 106 | 541 | -0.03 | 0.784 | -0.02 | 0.507 |
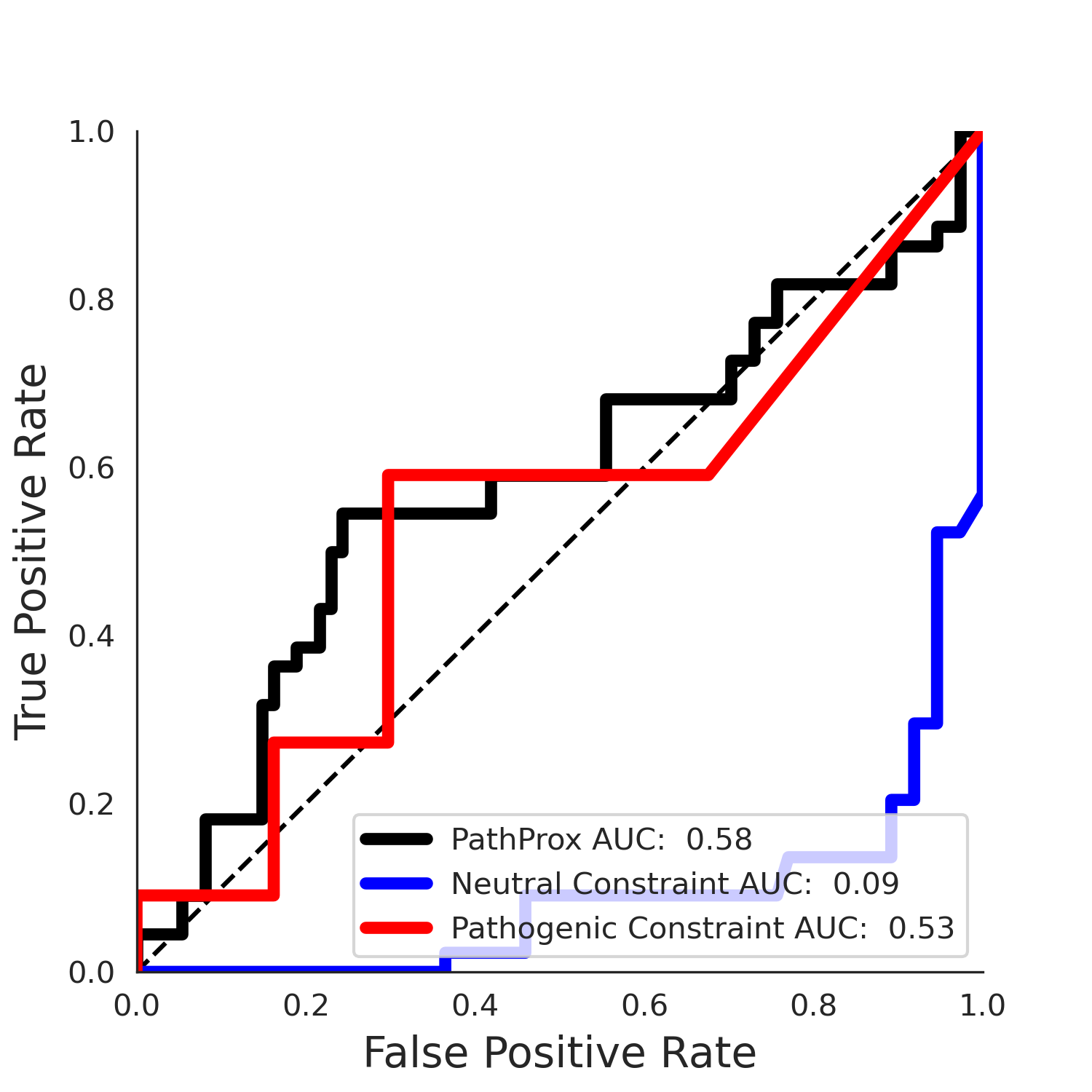
clinvar Results - 5qqr.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -3.68 | 36.00 | 1.97 | 15.00 | 0.83 | 0.78 | -0.03 | 0.98 | 0.01 |
COSMIC Results - 5qqr.A
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -3.33 | 34.00 | -1.58 | 11.00 | 0.53 | 0.51 | -0.02 | 0.99 | 0.00 |
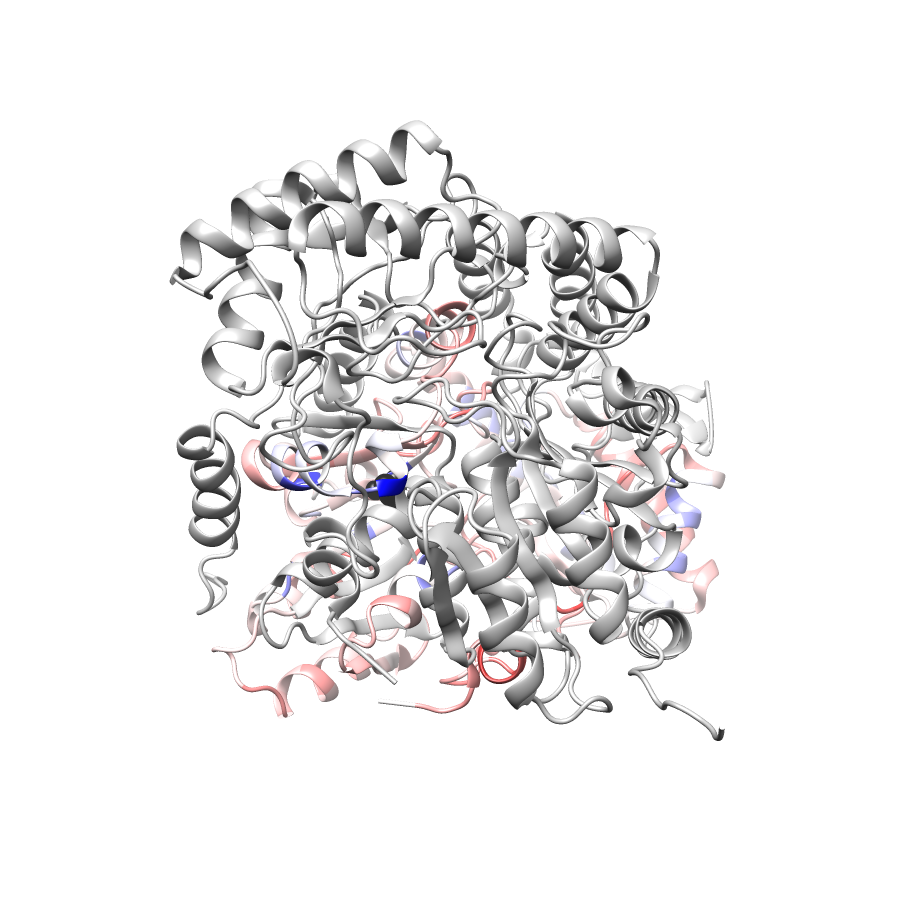
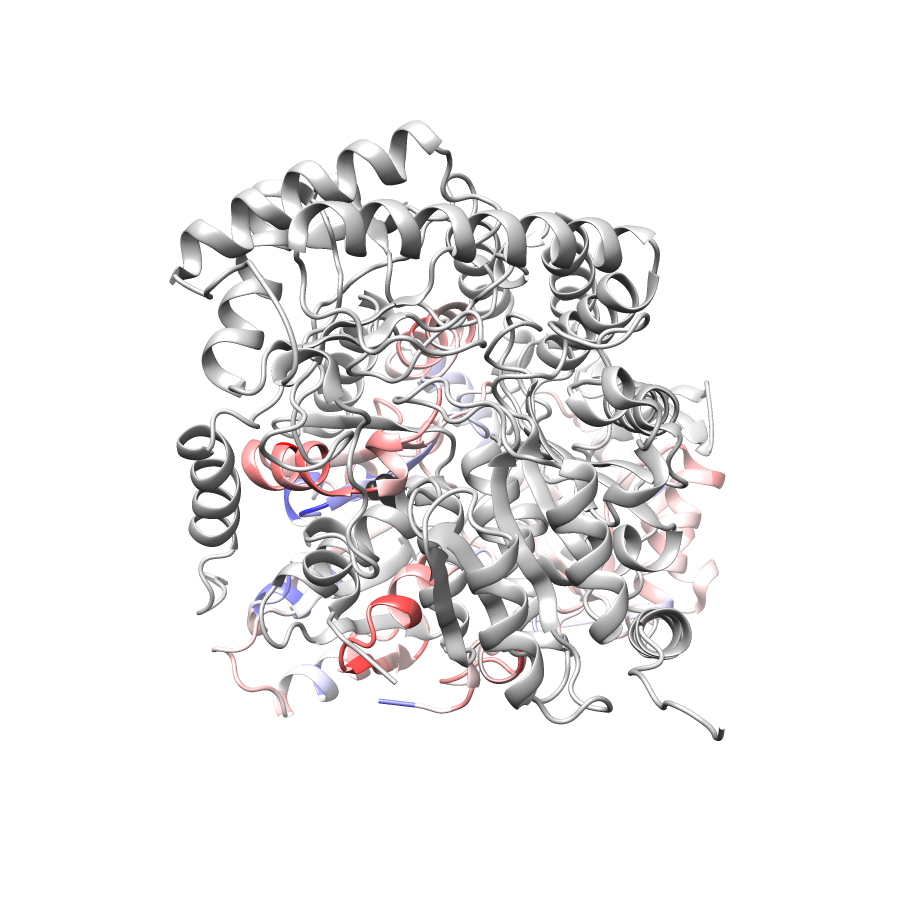
Location of A245T in 5qqr.A (NGL Viewer)
Variant Distributions
74 gnomAD38

46 clinvar
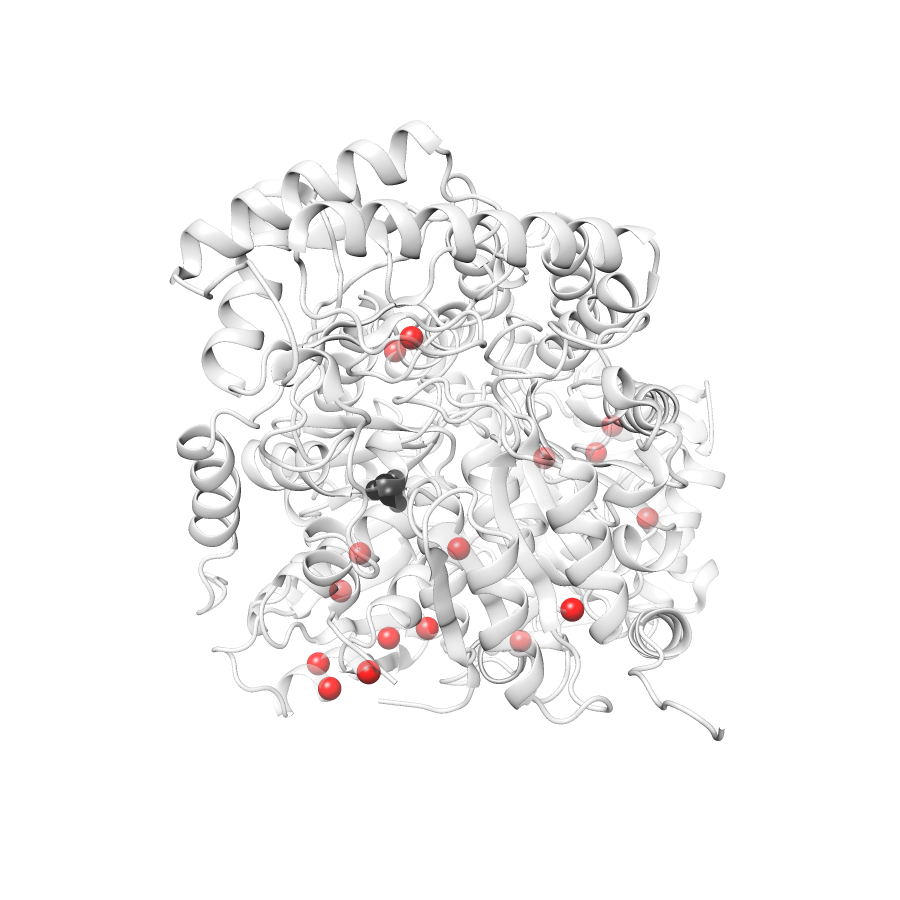
44 COSMIC
Ripley's Univariate K
gnomAD38
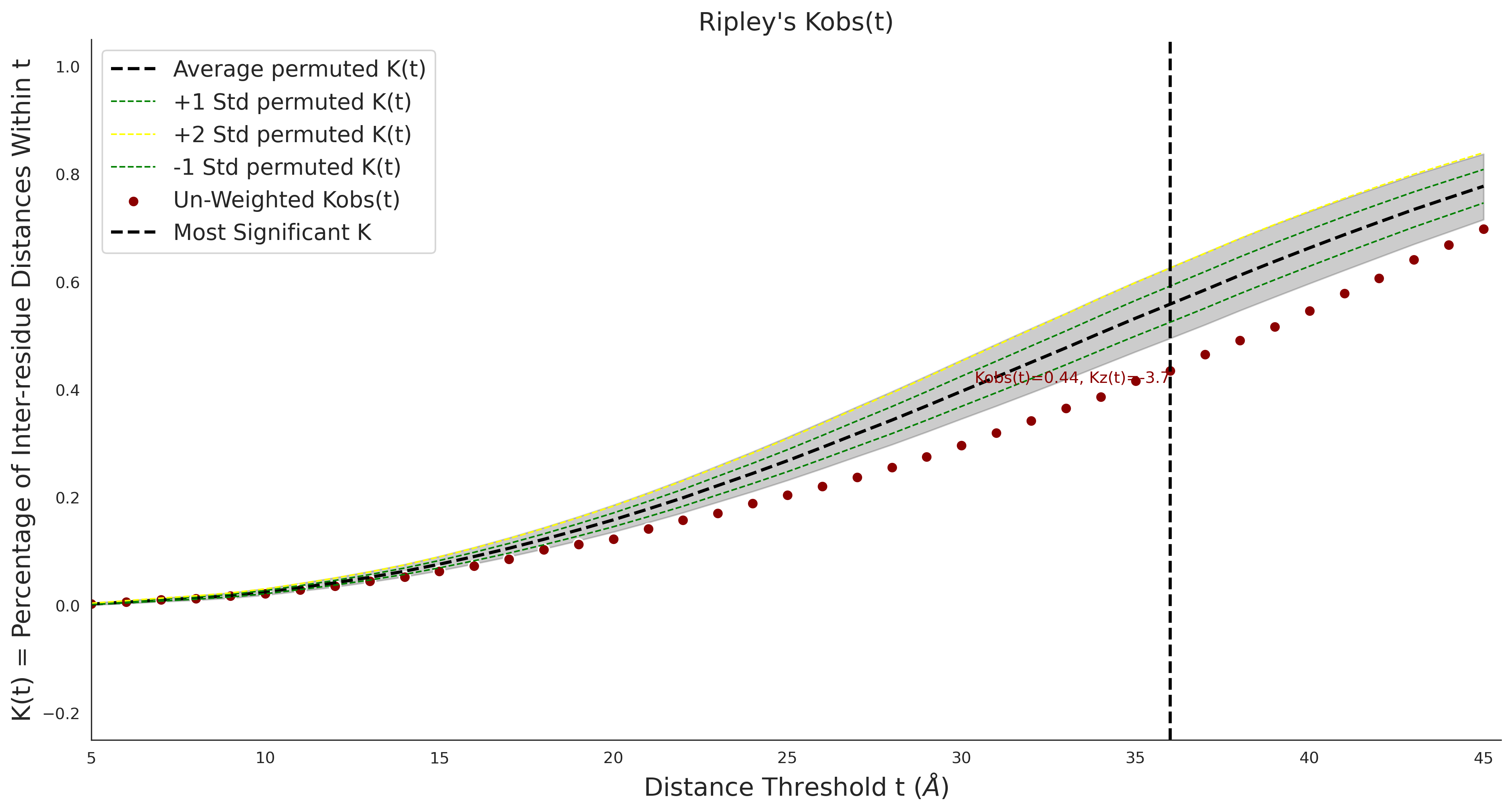
clinvar
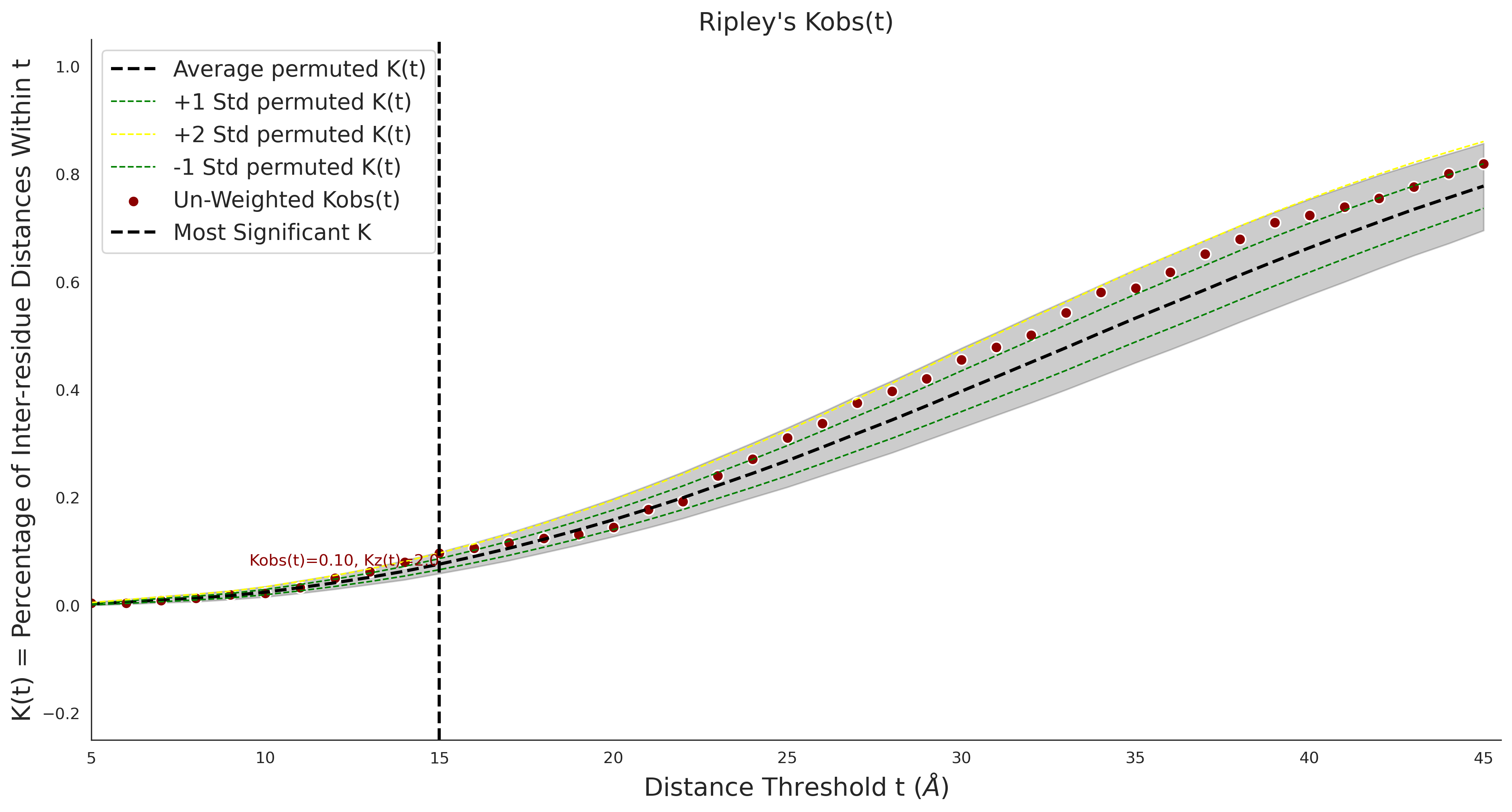
COSMIC
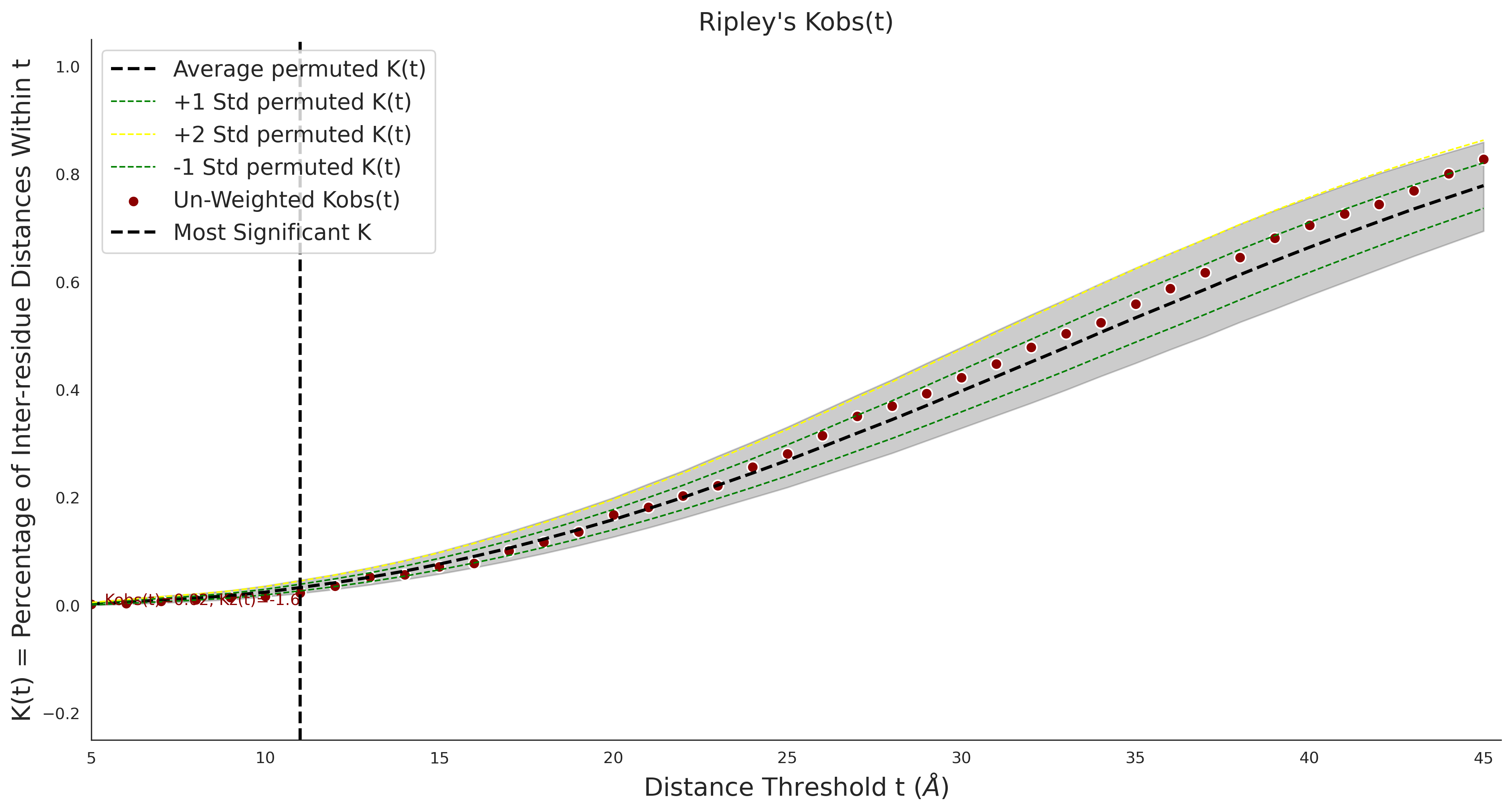
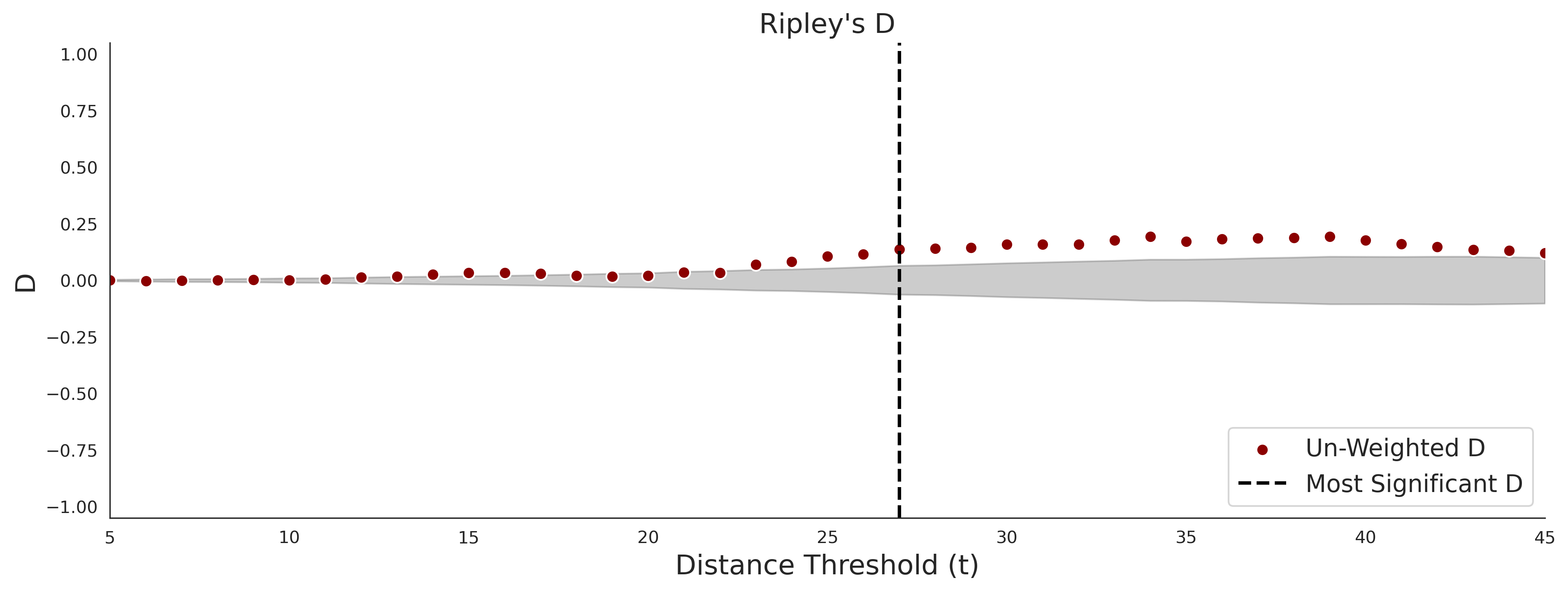
Ripley's Bivariate D
clinvar - gnomAD38
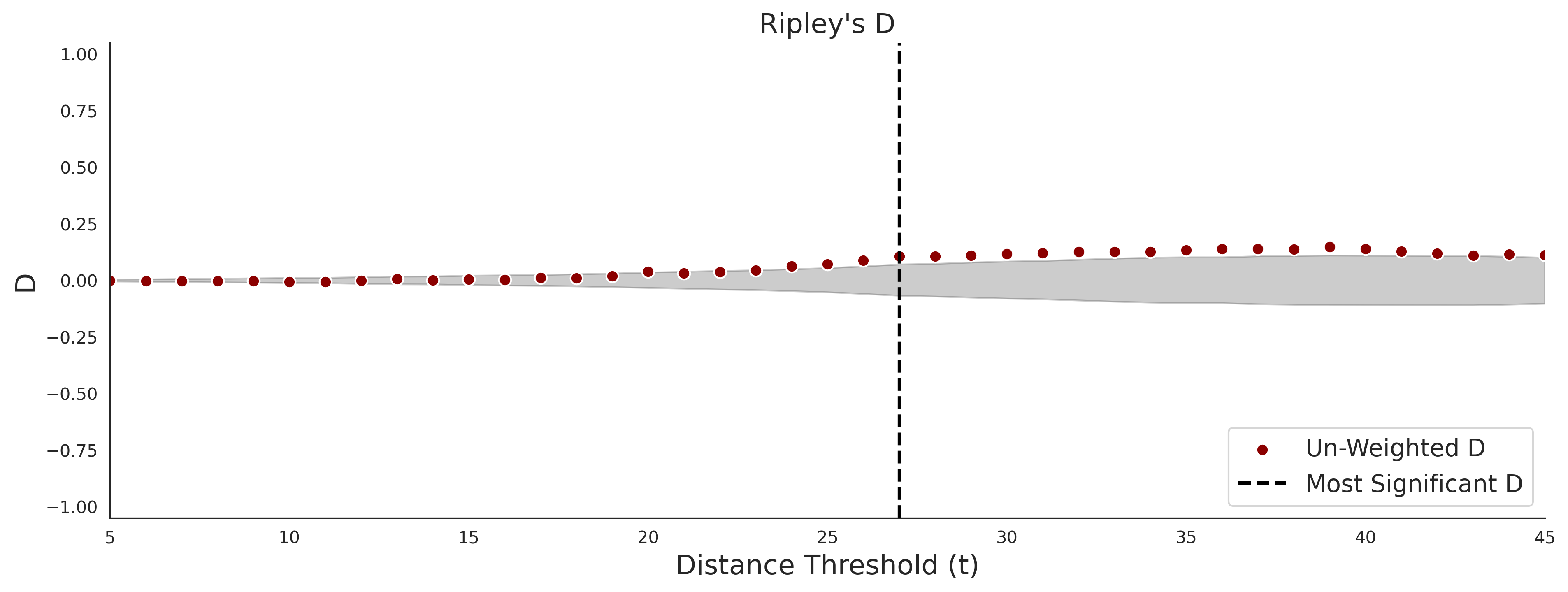
COSMIC - gnomAD38
PathProx (clinvar - gnomAD38) Results
PathProx
PathProx clinvar ROC
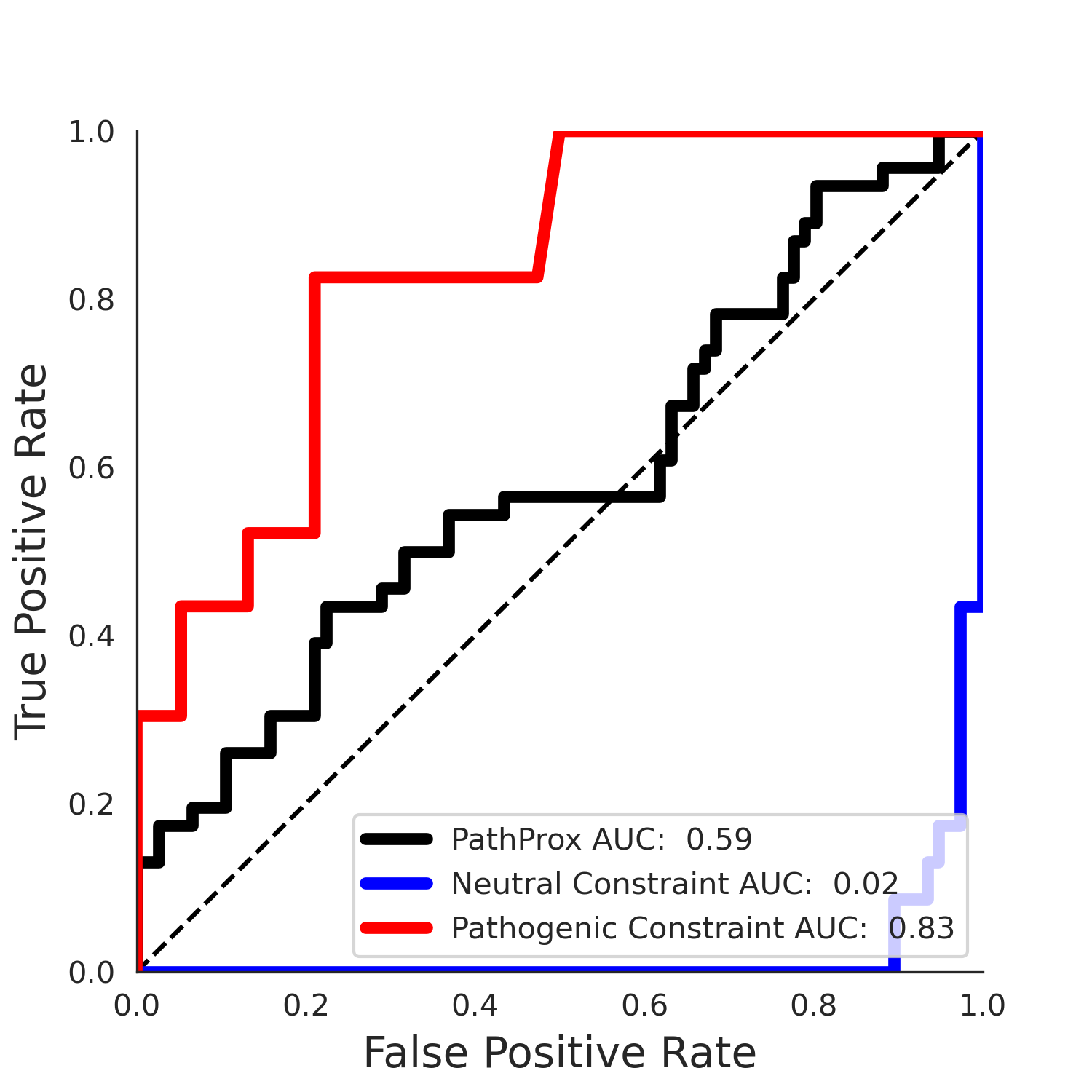
PathProx clinvar PR
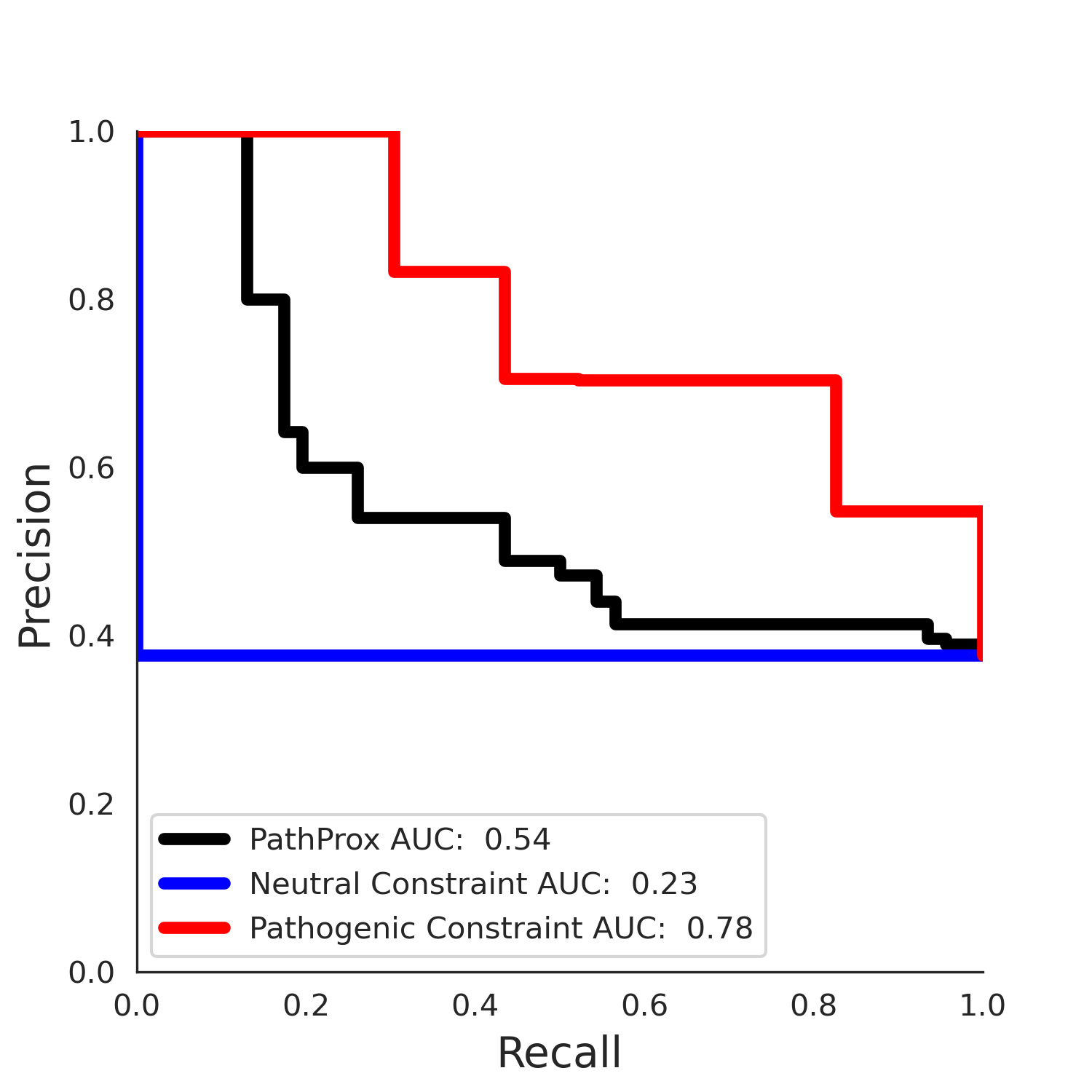
PathProx (COSMIC - gnomAD38) Results
PathProx
PathProx COSMIC ROC
PathProx COSMIC PR
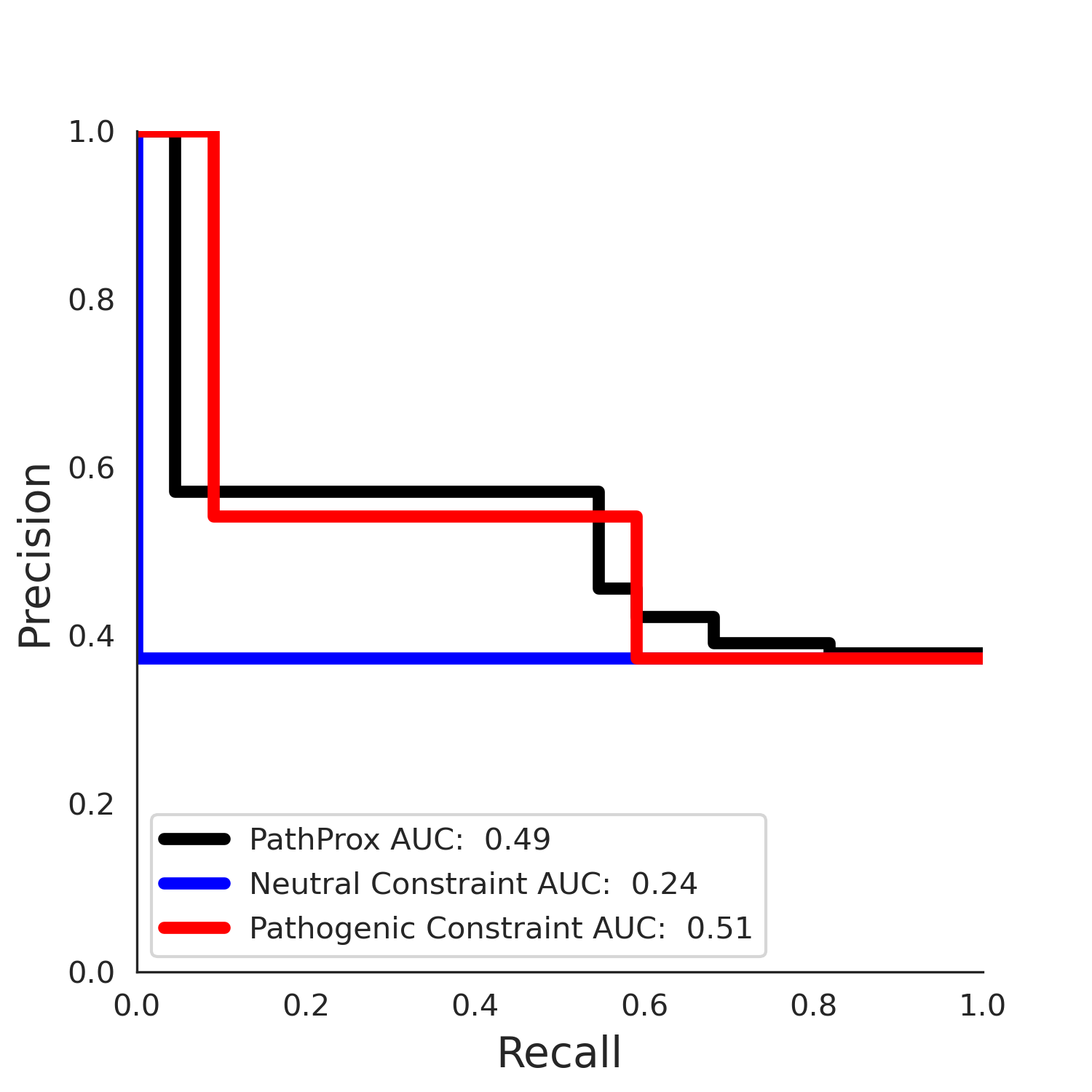
5qqr.B monomer - Structure Detail
| Method | Structure Id | Chain | PDB Template |
PDB Resolution |
Template Identity |
Transcript Identity |
AF Global Quality |
AF Residue Confidence |
Seq Start |
Seq End |
ddG Monomer |
ddG Cartesian |
Pathprox clinvar |
Pathprox clinvar AOC |
Pathprox COSMIC |
Pathprox COSMIC AOC |
PDB Pos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| X-RAY | 5qqr | B | 1.46 | 1.0 | 106 | 541 | 23.86 | 8.74 | 0.31 | 0.954 | -0.03 | 0.53 |
clinvar Results - 5qqr.B
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -2.55 | 38.00 | 2.40 | 33.00 | 0.96 | 0.95 | 0.31 | 0.88 | 0.22 |
COSMIC Results - 5qqr.B
| Neutral Kz | Neutral Kt | Pathogenic Kz | Pathogenic Kt | ROC AUC | PR AUC | PathProx | Neutral Constraint | Pathogenic Constraint |
|---|---|---|---|---|---|---|---|---|
| -2.50 | 36.00 | -1.20 | 11.00 | 0.54 | 0.53 | -0.03 | 0.99 | 0.00 |
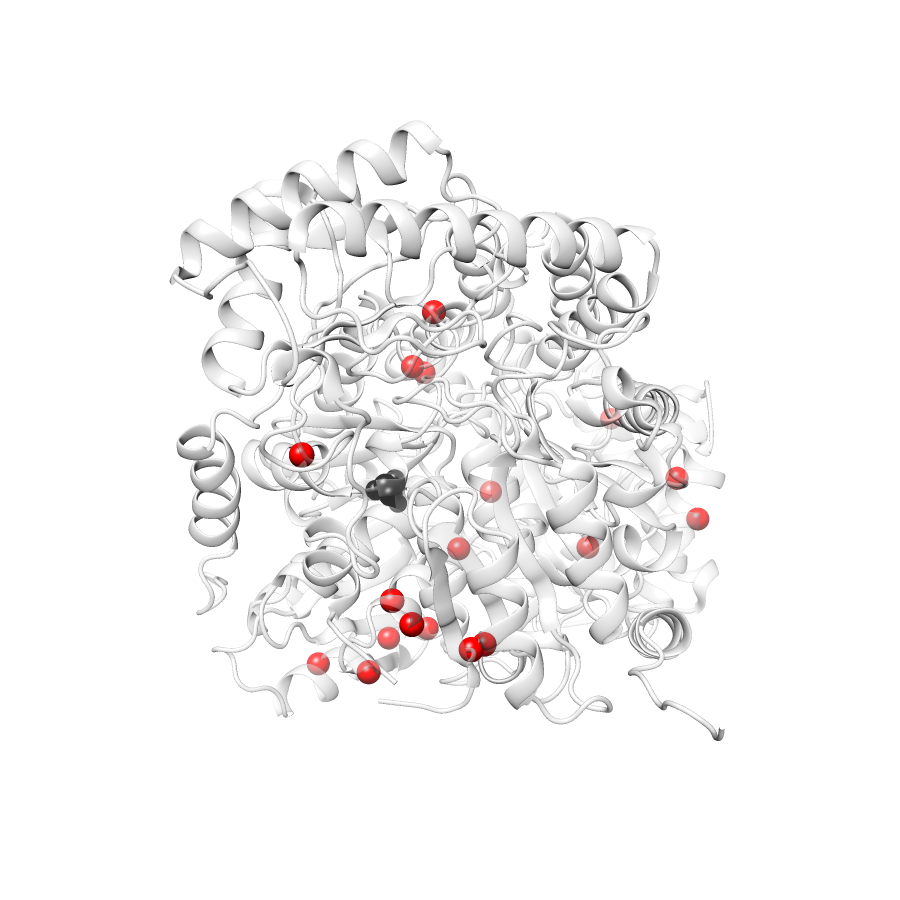
Location of A245T in 5qqr.B (NGL Viewer)
Variant Distributions
37 gnomAD38

23 clinvar

22 COSMIC

Ripley's Univariate K
gnomAD38
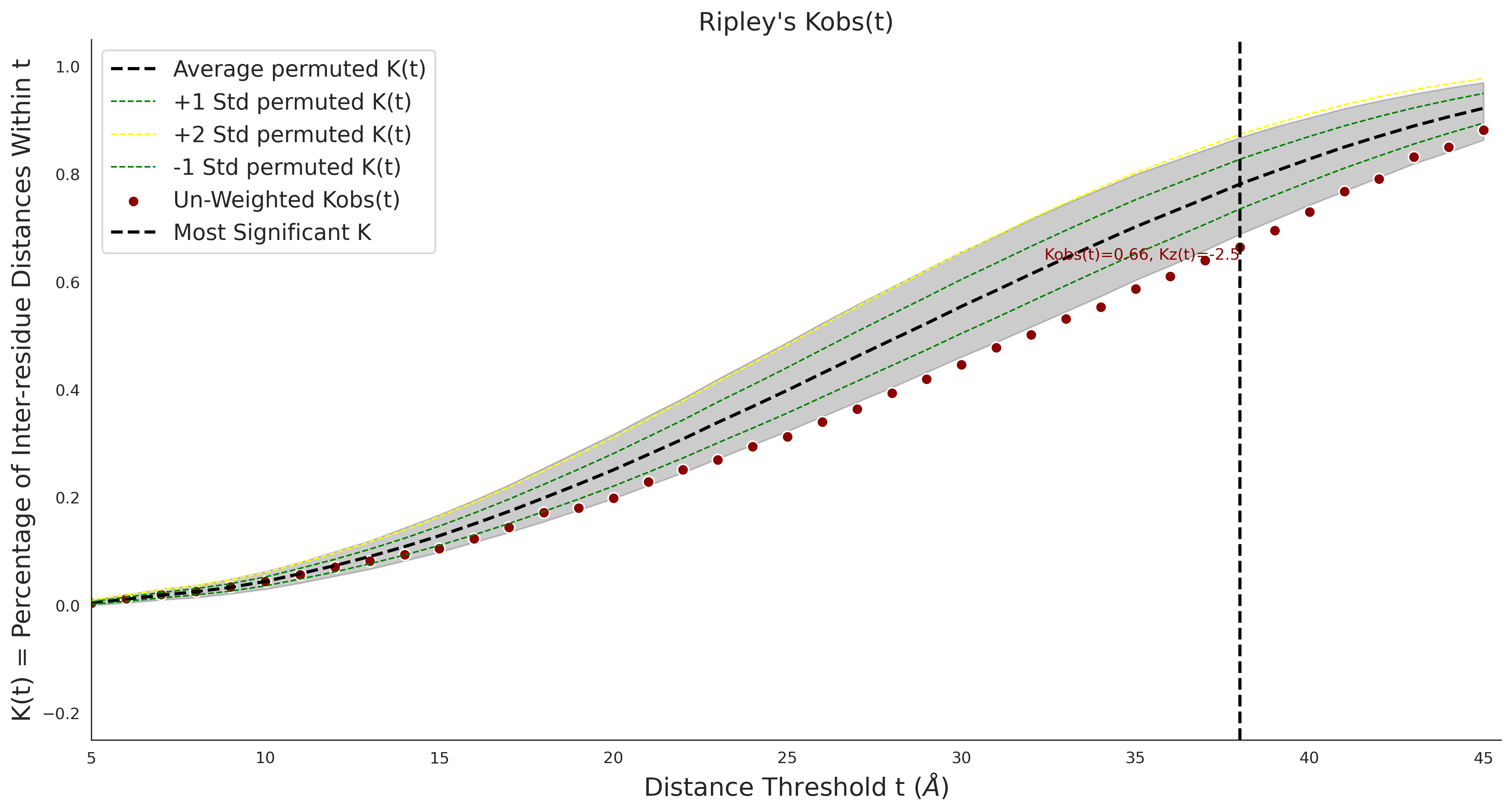
clinvar
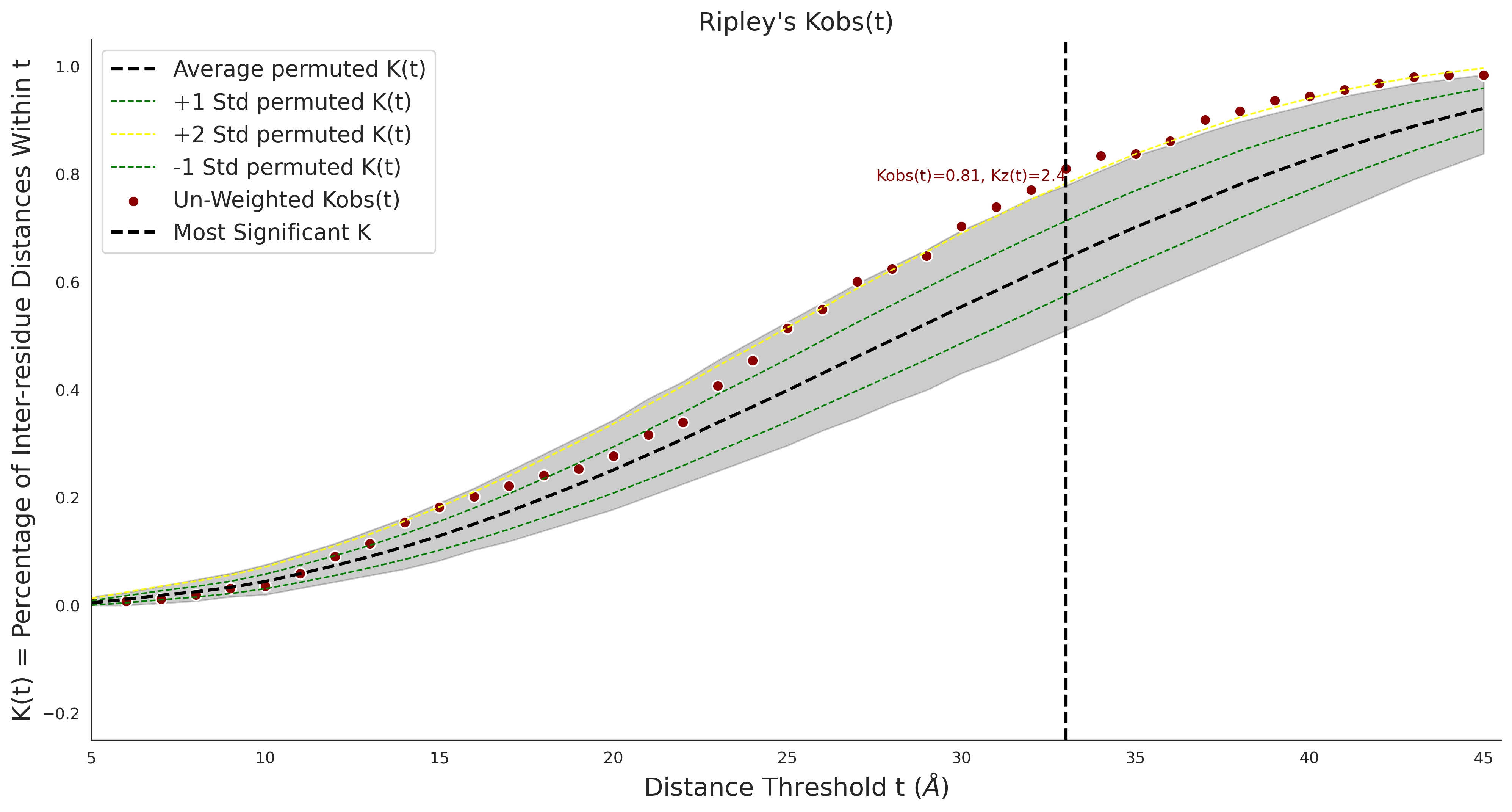
COSMIC
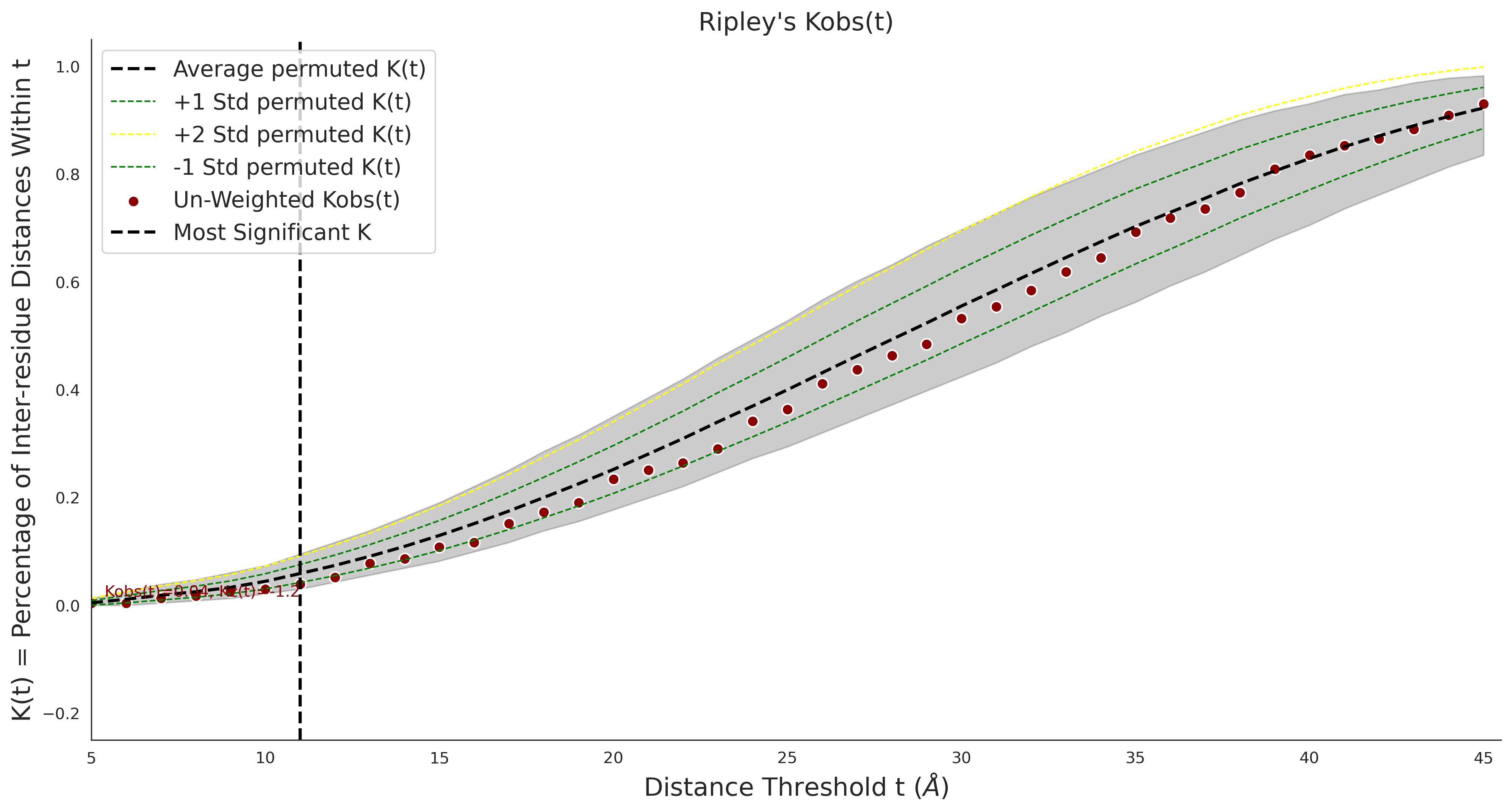
Ripley's Bivariate D
clinvar - gnomAD38
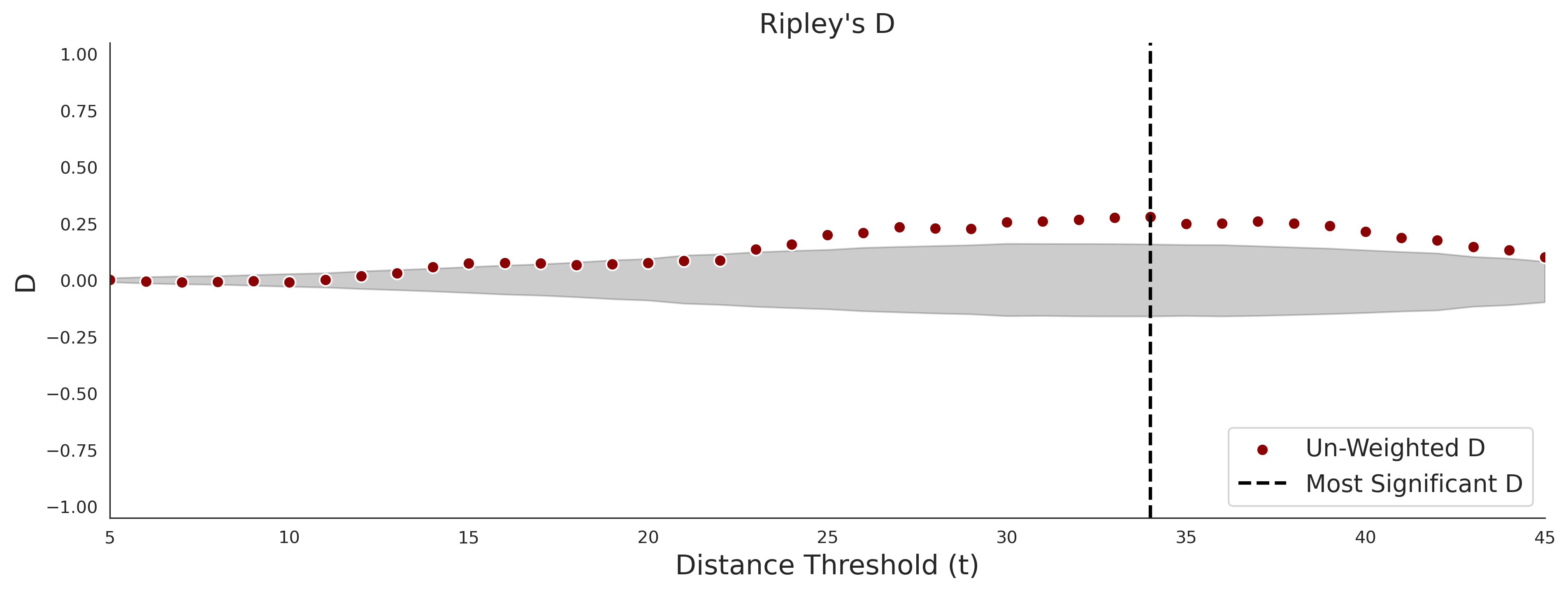
COSMIC - gnomAD38
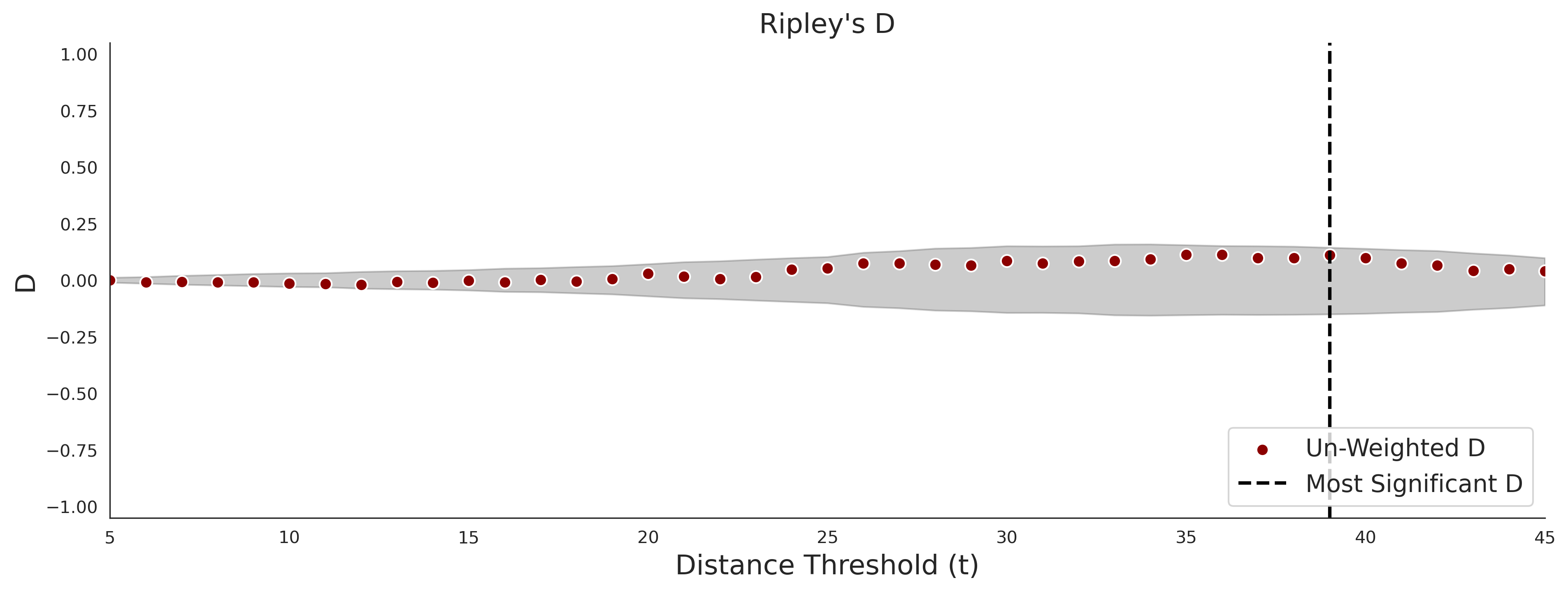
PathProx (clinvar - gnomAD38) Results
PathProx
No graphic was createdPathProx clinvar ROC
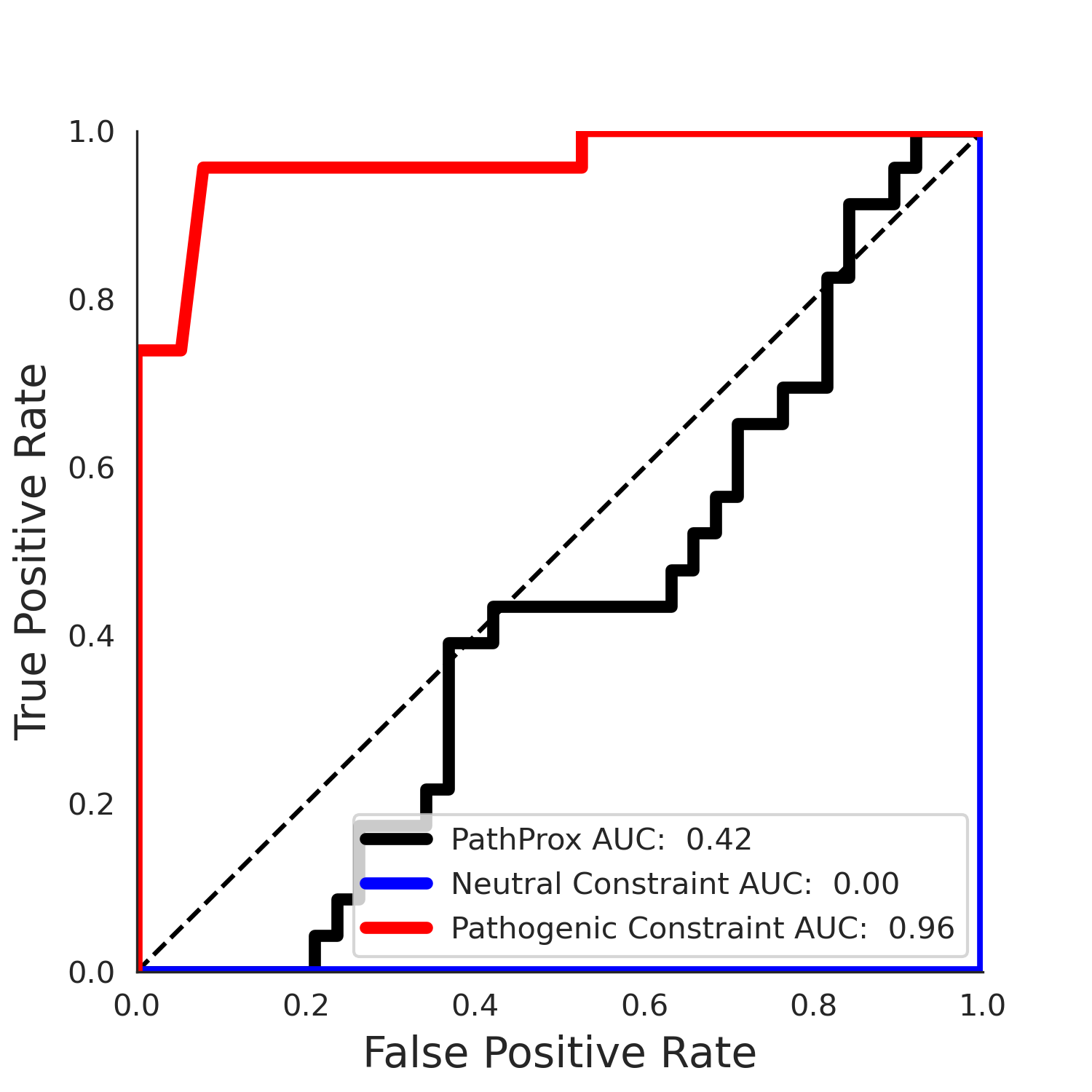
PathProx clinvar PR
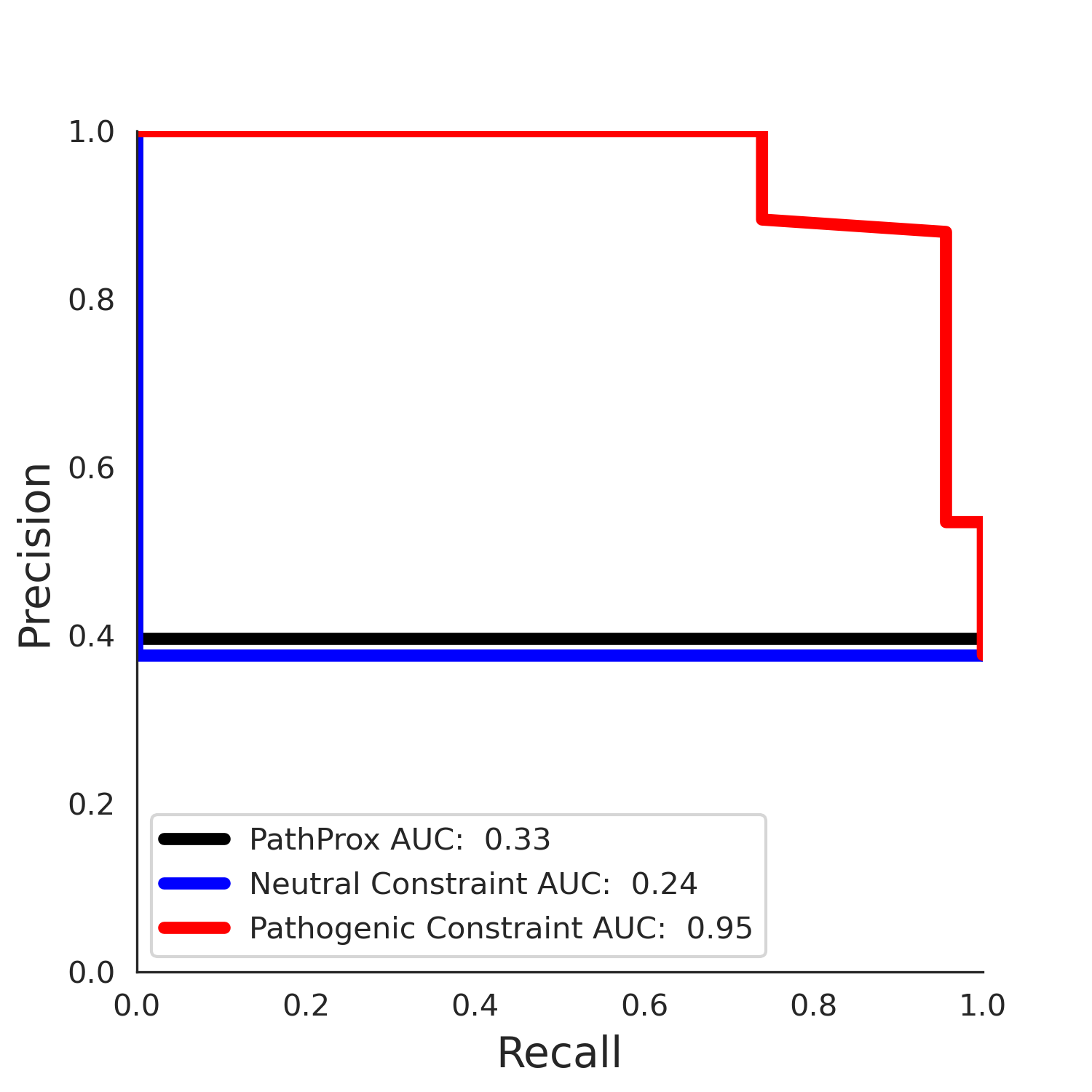
PathProx (COSMIC - gnomAD38) Results
PathProx
No graphic was createdPathProx COSMIC ROC
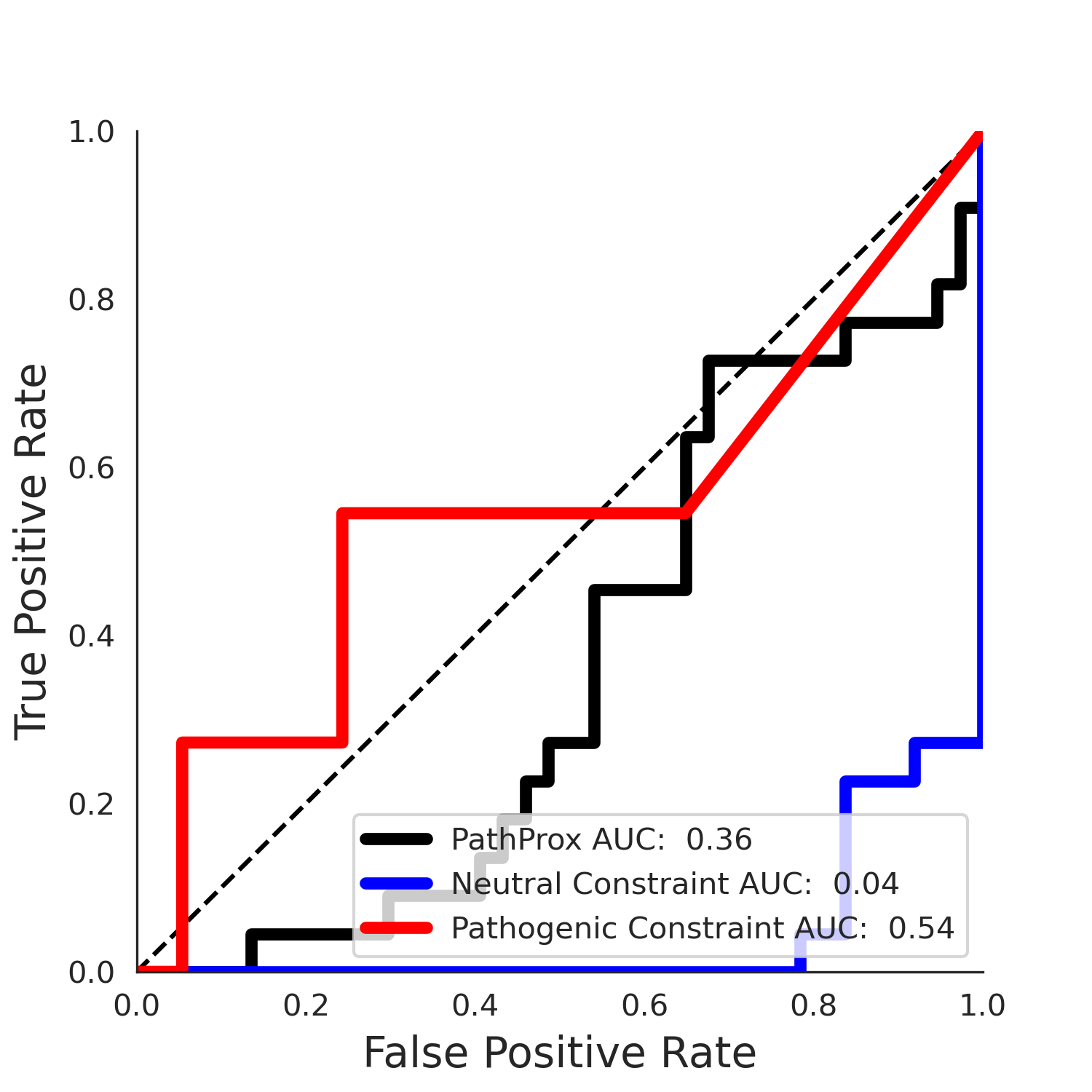
PathProx COSMIC PR