SampleCase
| # | Gene | Chrom | Pos | Variant | PP clinvar | PP COSMIC | ddg Monomer | ddg Cartesian | Gene Interactions | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Protein Unp ID |
Transcript Refseq |
Transcript ENSEMBL |
Prot Change |
Amino Acid Length |
Min | Max | Min | Max | Min | Max | Min | Max | Generic | Familial | Notes | |
| 0 | COA5 | chr2 | 98608380 | G/A | 74 | -0.39 | -0.39 | 2.37 | 2.37 | 3.35 | 3.35 | |||||
| Report | Q86WW8 | NM_001008215.2 | ENST00000328709.8 | P9L | 74 | -0.39 | -0.39 | 2.37 | 2.37 | 3.35 | 3.35 | |||||
| 1 | NAGA | chr22 | 42067209 | C/T | 411 | -0.01 | 0.11 | -2.36 | -0.66 | -0.08 | 0.78 | |||||
| Report | P17050 | NM_000262.2 | ENST00000396398.8... | D136N | 411 | -0.01 | 0.11 | -2.36 | -0.66 | -0.08 | 0.78 | |||||
| 2 | SMYD5 | chr2 | 73218934 | C/A | 418 | 0.01 | 0.05 | 27.1 | 27.1 | 7.34 | 7.34 | |||||
| Report | Q6GMV2 | NM_006062.2 | ENST00000389501.9 | A57E | 418 | 0.01 | 0.05 | 27.1 | 27.1 | 7.34 | 7.34 | |||||
| 3 | SMARCAL1 | chr2 | 216447069 | G/A | 954 | 0.06 | 0.47 | -0.03 | 0.31 | 0.67 | 4.43 | -0.13 | 10.3 | |||
| Report | Q9NZC9 | NM_001127207.1 | ENST00000357276.9... | A588T | 954 | 0.06 | 0.47 | -0.03 | 0.31 | 0.67 | 4.43 | -0.13 | 10.3 | |||
| 4 | ONECUT1 | chr15 | 52789080 | G/C | 465 | 0.04 | 0.04 | -2.92 | -2.92 | -1.97 | -1.97 | |||||
| Report | Q9UBC0 | NM_004498.2 | ENST00000305901.7 | R269G | 465 | 0.04 | 0.04 | -2.92 | -2.92 | -1.97 | -1.97 | |||||
| 5 | ALAS2 | chrX | 55017645 | C/T | 550-587 | -0.03 | 0.5 | -0.03 | 0.14 | 0.42 | 23.86 | 2.81 | 11.32 | |||
| Report | P22557-1 | NM_000032.4 | ENST00000650242.1 | A282T | 587 | 0.06 | 0.5 | -0.02 | 0.03 | 0.42 | 23.53 | 2.81 | 10.58 | |||
| Report | P22557-2 | NM_001037967.3 | ENST00000335854.8 | A245T | 550 | -0.03 | 0.5 | -0.03 | 0.03 | 2.3 | 23.86 | 2.97 | 11.32 | |||
| Report | P22557-4 | NM_001037968.3 | ENST00000396198.7 | A269T | 574 | 0.0 | 0.5 | -0.02 | 0.14 | 2.39 | 23.86 | 2.81 | 11.29 | |||
| 8 | AP3D1 | chr19 | 2115592 | C/T | 1153-1215 | -0.07 | 0.03 | -0.95 | -0.5 | -0.08 | 0.26 | |||||
| Report | O14617-1 | NM_003938.6 | ENST00000345016.9 | V699M | 1153 | -0.07 | -0.01 | -0.95 | -0.5 | -0.08 | 0.26 | |||||
| Report | O14617-5 | NM_001261826.1 | ENST00000643116.3 | V699M | 1215 | 0.03 | 0.03 | |||||||||
| 10 | RTEL1 | chr20 | 63688301 | G/A | 996-1300 | -0.01 | 0.49 | -0.03 | 0.12 | 2.03 | 2.03 | 0.12 | 0.12 | |||
| Report | Q9NZ71-1 | NM_016434.3 | ENST00000370018.7 | G546D | 1219 | 0.0 | 0.46 | -0.03 | 0.12 | 2.03 | 2.03 | 0.12 | 0.12 | |||
| Report | Q9NZ71-6 | NM_001283009.1 | ENST00000360203.11 | G546D | 1300 | -0.01 | -0.01 | -0.01 | -0.01 | |||||||
| Report | Q9NZ71-7 | NM_032957.4 | ENST00000508582.7 | G570D | 1243 | 0.16 | 0.49 | -0.01 | -0.01 | |||||||
| Report | Q9NZ71-8 | NA | ENST00000482936.6 | G546D | 1004 | |||||||||||
| Report | Q9NZ71-9 | NM_001283010.1 | ENST00000318100.9 | G323D | 996 | 0.23 | 0.35 | -0.02 | 0.01 | |||||||
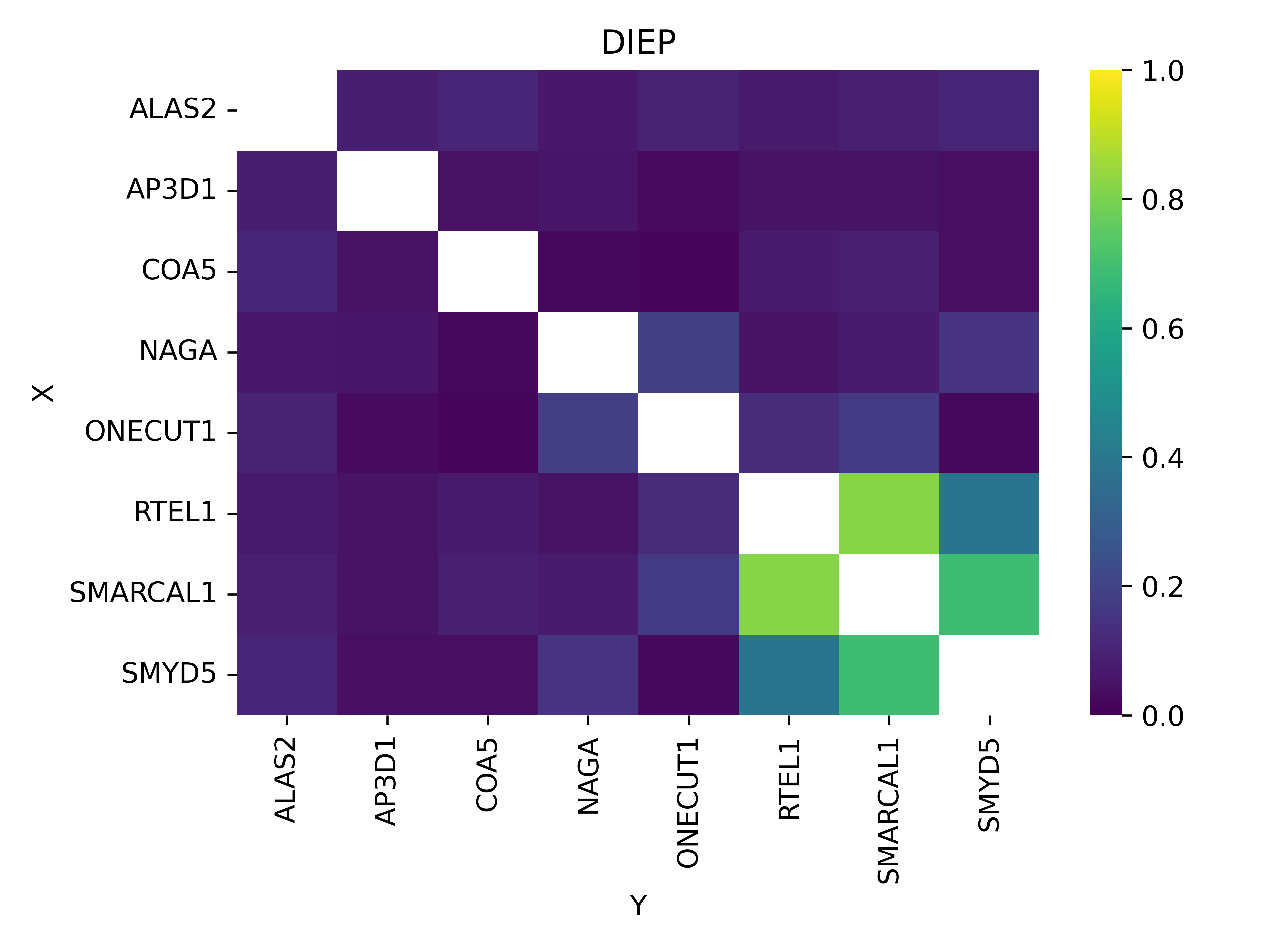
DiGePred Analysis
Top Scoring DiGePred Gene Pairs
| DiGePred Score | Gene A | Gene B |
|---|---|---|
| 0.547 | COA5 | NAGA |
| 0.166 | AP3D1 | NAGA |
| 0.14 | COA5 | RTEL1 |
| 0.0762 | ALAS2 | COA5 |
| 0.0759 | COA5 | SMARCAL1 |
| 0.0753 | NAGA | SMARCAL1 |
| 0.0531 | NAGA | RTEL1 |
| 0.0368 | RTEL1 | SMARCAL1 |
| 0.019 | RTEL1 | SMYD5 |
| 0.0182 | ALAS2 | RTEL1 |
| 0.0166 | COA5 | ONECUT1 |
| 0.00898 | AP3D1 | SMARCAL1 |
| 0.00824 | NAGA | ONECUT1 |
| 0.00626 | ONECUT1 | RTEL1 |
| 0.00603 | ONECUT1 | SMARCAL1 |
| 0.00435 | AP3D1 | RTEL1 |
| 0.00434 | ALAS2 | ONECUT1 |
| 0.00316 | NAGA | SMYD5 |
| 0.00203 | ALAS2 | SMARCAL1 |
| 0.000479 | AP3D1 | COA5 |
DIEP Digenic Interaction Analysis Heatmap